Poly-histidine tagged proteins account for more than 90% of routine recombinant protein expression and purification. Histidine tag has many advantages over other affinity tags such as small size, high affinity for binding to IMAC resins (Immobilized metal affinity chromatography) and allows one step purification of recombinant protein. But sometimes a histidine tagged protein doesn’t bind to the affinity column and evades it’s capture and subsequent purification. Although such instances are not very common but can be very frustrating whenever they happen. However, there are ways to troubleshoot if such a problem arises and the solutions are discussed below in a step by step manner.
Does your solubilisation buffer need to be optimized?
Ni-NTA resins are sensitive to presence of reducing agents such as DTT and chelating agents such as EDTA or EGTA, make sure that the solubilisation buffers do not contain any of these substances, since they affect the binding capacity of the IMAC resins.
Usually an Imidiazole concentration of 15-25mM is maintained in the protein solubilisation/binding buffer to prevent binding of non-specific proteins to the IMAC resins, It is recommended to check the imidiazole concentration in solubilisation buffer, sometimes even low imidiazole concentrations can prevent binding of proteins to the Ni-NTA resin. In such one should carefully try lower concentrations that don’t interfere with the binding of the protein with affinity resin. Another critical factor is pH of the solubilisation buffer. When the pH of the solubilisation buffer is low, it can cause protonation of the histidine side chain and this can prevent histidine residues from coordinating with the metal required for successful binding. It is usually recommended to keep the pH of the buffer close to the isoelectric point of the protein. Addition of Imidiazole can lower the pH and it is therefore recommended to adjust the pH of the binding/solubilisation buffer after the addition of imidazole.
Is your affinity resin fresh and working ?
To ensure the affinity resin is working, perform batch binding of a control his-tagged protein (one that has been purified earlier using IMAC) along with your protein of interest both in same buffer. If control protein binds and elutes with imidiazole but your protein of interest continue to appear in flow through and column washes and doesn’t elute, it is time to get serious and look for other causes.
Always use fresh Ni-NTA when trying to purify a novel protein for the first time.
Is your protein hiding the histidine tag ?
Certain proteins can hide the polyhistidine tag in their folded native structure and thus prevents the binding of the protein to the IMAC affinity column. The result is that tag is not accessible to coordinate with the metal, typically nickel or cobalt. Inaccessibility of the tag can be due to tag being buried within three-dimensional conformation of a folded protein. A quick and reliable way to find this out is to perform the purification in the presence of strong denaturants such as Urea or Guanidinium Chloride in the solubilisation buffer. Both urea and guanidinium chloride are powerful denaturants and would unfold the protein, if the protein binds to the resin in presence of these denaturants, it can be said with certainty that folded protein hides the his-tag and prevents its binding to the IMAC resin. 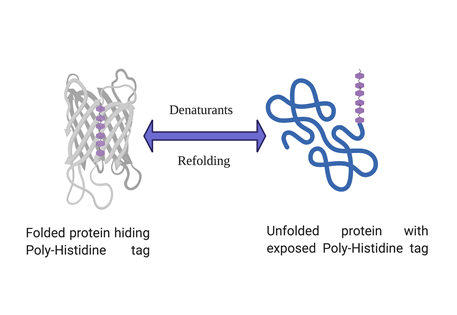
How to overcome it ?
There are many ways to overcome the problem of a hidden histidine tag, some of them are as follows
- Purify the protein under denaturing conditions and refold it later on.
- Add a linker sequence to separate the protein from the poly-histidine tag from the protein, usually a linker of poly-serine or poly-glycine can be used. This strategy calls for intervention at the cloning step. The presence of a flexible linker ensure that the polyhistidine tag remains available for binding to IMAC resin and is not hidden by the protein.
- Final option is to place the poly-His tag to the opposite terminus and see if such a swap allows the His-tag to bind to the affinity matrix.
Usually one of the these strategies will work but in case they still do not work, then one can go for use of larger affinity tag such as GST or MBP instead of small Histidine tag.





