The cloning and expression of proteins to product recombinant proteins is a hugely popular technique that allows for the production of copious quantities of protein that almost mimic native proteins. Although a popular and useful technique, one of the main reasons for its use can also be a drawback. In some cases, over-expression of the recombinant protein leads to an accumulation of misfolding protein that aggregates and is sequestered into inclusion bodies. The purification of inclusion bodies is time consuming, involves harsh denaturants, detergents and other chemicals, resulting in damaged and denatured proteins. If a researcher is successful in extracting these proteins then they must invest more time and effort to refold the denatured proteins. For this reason, researcher's want to prevent their recombinant proteins from ending up in inclusion bodies.
Cold-Shock Keeps Recombinant Proteins out of Inclusion Bodies
Topics: Protein Extraction
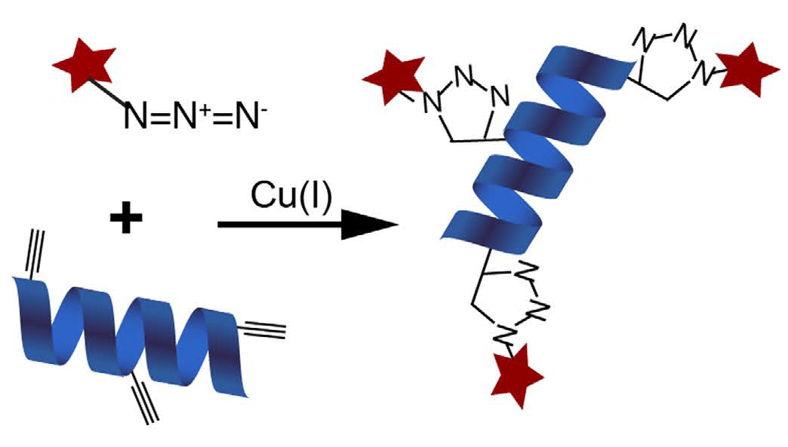
BONCAT (Bioorthogonal Non-canonical Amino Acid Tagging): a method to define the translatome
Living entities have a tightly regulated dynamic system, where majority of the energy expenditure goes for maintaining the homeostasis. Most of the biological processes such as growth, development, metabolism etc are regulated at the crucial control point of protein translation and degradation (rate of protein turnover). Studies on different organisms/model systems have suggested that transcriptome modifies itself dynamically in response to different cellular conditions. The translational regulation is a slightly independent add-on layer of control after that, hence, during the cell processes a linear relationship of transcriptome and translatome is relatively difficult to establish. The efficacious identification of lesser abundant proteins is of lower magnitude, albeit of having sub-femtomolar level of sensitivity of modern state-of-the- art Mass spectroscopic instruments because of a narrow dynamicity and sequencing speed.
Topics: Protein Labeling, Mass Spectrometry
Aside from tissue biopsy, cytology serves as an indispensable tool in screening and diagnosing cancer. In this technique, a cytological material is obtained from the patient, spread onto glass slides for microscopic examination, stained, screened for abnormalities and assessed prior to the issuance of a final report. Cytology differs from histology in that the sample usually consists of a suspension of cells whereas histology samples are usually sections of actual tissue. For example; a Gill’s hematoxylin single strength formulation would be much better suited for cytology, whereas a triple strength formulation would be better for tissue sections.
Topics: Cytotoxicity Assays
SILAC: A General Workflow for Improved Mass Spectrometry
This is a follow up blog to SILAC for Improved Mass Spectrometry & Quantitative Proteomics
SILAC (Stable Isotope Labeling with Amino Acids in Cell Culture) method of working is differentiated into two critical phases: an adaptation phase and an experimental phase. Before initiating the adaptation phase, it is crucial to characterize the cell type to be used for the labeling. In general, dialyzed serum is used to rear the cells in order to negate the availability of free amino acids present in the normal serum. Although, some cell lines do not grow that well in a dialyzed medium due to the absence of some of the growth factors, therefore, supplementation of purified growth factors with the dialyzed media or small percentage of normal serum in the dialyzed media may be helpful.
Topics: Mass Spectrometry