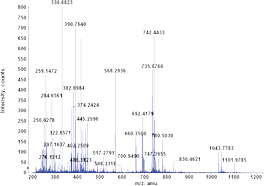
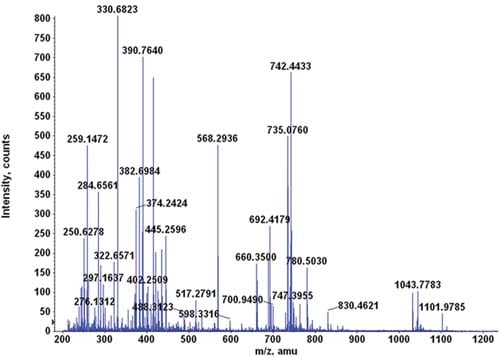
Fragmentation occurs when energetically unstable molecular atoms dissociate (either through homolytic or heterolytic cleavage) as they pass through the ionization chamber of a mass spectrometer. Here, the sample is bombarded with a stream of electrons powerful enough to remove an electron from the sample and form a positive ion (also called molecular or parent ion).
Spotting Fragmentation Patterns When Using Mass Spectrometry
Topics: Mass Spectrometry
The Advantages of Using Trypsin for Mass Spectrometry
In mass spectrometry-based proteomics, nothing comes close to trypsin in breaking down protein mixtures into peptide fragments. In fact, protein researchers consider trypsin as the runaway winner when it comes to protease activity and specificity – and there are a lot of good reasons why they do.
Topics: Mass Spectrometry
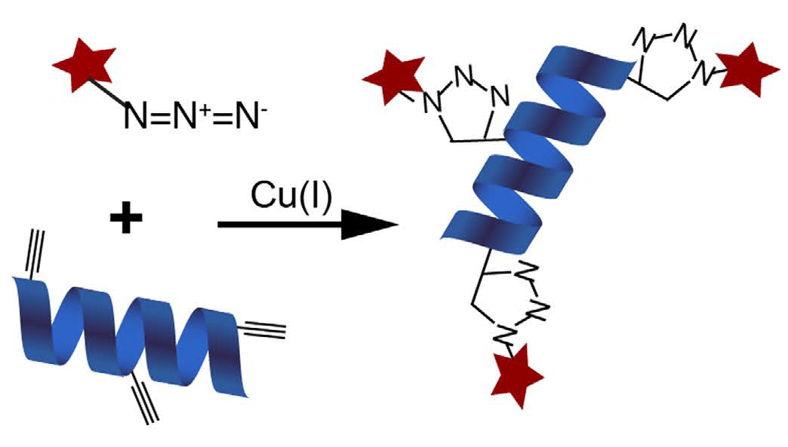
BONCAT (Bioorthogonal Non-canonical Amino Acid Tagging): a method to define the translatome
Living entities have a tightly regulated dynamic system, where majority of the energy expenditure goes for maintaining the homeostasis. Most of the biological processes such as growth, development, metabolism etc are regulated at the crucial control point of protein translation and degradation (rate of protein turnover). Studies on different organisms/model systems have suggested that transcriptome modifies itself dynamically in response to different cellular conditions. The translational regulation is a slightly independent add-on layer of control after that, hence, during the cell processes a linear relationship of transcriptome and translatome is relatively difficult to establish. The efficacious identification of lesser abundant proteins is of lower magnitude, albeit of having sub-femtomolar level of sensitivity of modern state-of-the- art Mass spectroscopic instruments because of a narrow dynamicity and sequencing speed.
Topics: Protein Labeling, Mass Spectrometry
SILAC: A General Workflow for Improved Mass Spectrometry
This is a follow up blog to SILAC for Improved Mass Spectrometry & Quantitative Proteomics
SILAC (Stable Isotope Labeling with Amino Acids in Cell Culture) method of working is differentiated into two critical phases: an adaptation phase and an experimental phase. Before initiating the adaptation phase, it is crucial to characterize the cell type to be used for the labeling. In general, dialyzed serum is used to rear the cells in order to negate the availability of free amino acids present in the normal serum. Although, some cell lines do not grow that well in a dialyzed medium due to the absence of some of the growth factors, therefore, supplementation of purified growth factors with the dialyzed media or small percentage of normal serum in the dialyzed media may be helpful.
Topics: Mass Spectrometry