A restriction enzyme (restriction endonuclease) is a special enzyme that recognizes a specific sequence of nucleotides and cleaves DNA at that specific site (restriction site or target sequence). These enzymes, which are usually found in bacteria and other prokaryotes, are considered as one of the most important tools in recombinant DNA technology since they can easily cut DNA into fragments and/or join DNA molecules from different genomes so researchers can identify and characterize genes and examine gene expression and regulation.
The Protein Man

Recent Posts
Restriction Enzyme Analysis: How to Make the Cut
Topics: Molecular Biology
Proteomic Grade Detergents: Why you should use them
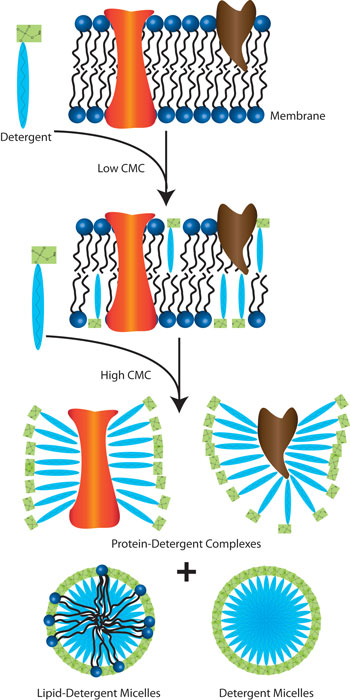
Detergents play an extremely critical role in cell lysis and protein extraction. With their unique amphipathic nature (they possess a polar head group on one end that interacts with the hydrogen bonds of water molecules and a long hydrophobic carbon tail which aggregate to form micelles on the other end), this class of molecules can easily disrupt the hydrophobic-hydrophilic interactions between biological molecules in aqueous solutions.
Topics: Detergents
Exosome isolation by centrifugation, filtration, immunoaffinity & more
Earlier designated as the "garbage bags" used by the biological systems to remove unnecessary biomolecules of the cells, Exosomes, have recently gained popularity as secreted vesicles pivotal for intracellular and intercellular information transfer. Studies have shown the presence of essential RNA and protein cargos in these small vesicles (30-150 nm). The interest in exosome studies have exponentially grown as they can be manoeuvred as minimally diagnostic tools in order to understand biological functions. Proteomics of exosomes are considered the biological fingerprints as they replicate the properties of the parental cell from where they are originated. As a mobile container of vital biomolecules (specialized proteins and RNAs) exosomes are crucial for antigen- presentation, cell-cell communication, waste management, translocation of biomolecules and coagulation.
Topics: Protein Extraction
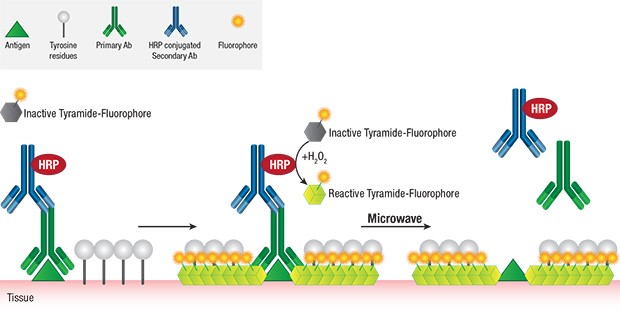
Diagnostic field has been a progressing area with multiple tools identified for specific proteins detection and their quantification. Diagnosis in pathology or clinical set up has multiple numbers of challenges owing to time constraint, increasing numerosity and complexity of the detection methods, less amount of sample and so on, which is essentially important for disease identification, refining of pathological interventions and development of an appropriate prognosis. Pathological detection is most commonly performed by utilizing antibody-antigen interactions (by deposition of a visible product or fluorochrome based detection). The standard approach is to perform one or two protein detection at a time on the serial sections, which is cumbersome if the sample availability is low and the proteins to be detected are high in number. More often fluorescent reporters are used for both detection and quantitation methods such as image cytometry, flow cytometry, confocal microscopy, etc. using live tissue or slightly fixed tissue, while the conventional immunohistochemistry involves heavily fixed tissue sections. Recently, in the wake of developing user-friendly and multiple assays on formalin fixed, paraffin embedded (FFPE) specimens are being developed and gaining ground, known as Multiplexing or multiplex immunohistochemistry (mIHC), which allows use of more than three different stains performed on one sample/ slide.
Topics: Assay Development (ELISA)