Evolution of ancient cellular components paves a path to parasitic adaptations in Apicomplexan parasites.
Parasites are everywhere! They co-exist with host and exploit host to ensure their survival. Apicomplexan parasites are considered model organisms to study the biology of cellular parasitism. They have evolved for such a long time for a parasitic mode of life that they know no other means to survive and proliferate, concisely put, they are obligate intracellular parasites. Apicomplexan parasites have a polarized cell structure, the apical end of the cell is adapted for host cell invasion, the name Apicomplexa signifies the presence of an apical complex of invasive organelles.
Apicomplexan parasites consist of well-known pathogens of human and veterinary importance such as Plasmodium, Babesia, Cryptosporidium, Eimeria and Toxoplasma gondi. Toxoplasma gondi is very peculiar amongst all, it has an incredible ability to infect any nucleated cell in a warm-blooded organism. What makes Toxoplasma such a successful parasite? A closer look at the cellular architecture reveals that Toxoplasma is highly adapted to invade or penetrate cells. Invasive cells of tachyzoites have a very sophisticated structure at the apical end known as conoid. A conoid is the linchpin of the Toxoplasma’s unusual parasitic prowess. A conoid is not only required for invasion but also for exit or escape from the host cell where the parasite has completed its multiplication.
“A conoid is the linchpin of the Toxoplasma’s unusual parasitic prowess. It is not only required for invasion but also for the exit or escape from the host cell where the parasite has completed its multiplication. Evolutionary biologists believe that conoid is a modified cilium that underwent evolutionary changes to suit a parasitic mode of lifestyle”.
Electron micrograph of a Conoid Extracellular forms of Toxoplasma


Image Source: https://www.ft.dk/samling/20151/almdel/MOF/bilag/205/1587343.pdf
Evolutionary biologists believe that conoid is a modified cilium that underwent evolutionary changes to suit a parasitic mode of lifestyle. Cilia and conoid have similar 9+2 microtubule arrangement which is also present in flagella. A cilium is an ancient cellular structure and is believed to have been present in the last common eukaryotic ancestor cell. Cilium is present in all eukaryotes but a conoid is found only in parasitic apicomplexans such as Toxoplasma. It is sensitive to calcium and can protrude in response to a local rise in calcium concentration. Conoid protrusion is necessary for invasion, it is required for secretion of proteins from secretory organelles present within the apical end of the parasite. The secretion of these proteins facilitates host cell invasion. The basic organization of cilia and conoid is similar and recent researches have further demonstrated their evolutionary relationship by a common pathway of biogenesis.
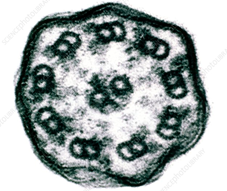
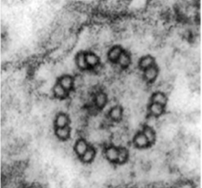
Cross section through a Cilium Cross section through a Conoid
The biogenesis or assembly of cilia requires the activity of ERK7, an ancient MAP kinase present in almost all eukaryotes. Surprisingly, it has been recently discovered that ERK7 activity plays a critical role in the biogenesis of the conoid in Toxoplasma (3). Abrogation of ERK7 results in complete loss of Toxoplasma’s ability to invade host cells or even egress from the cells once the multiplication is complete. Electron microscopic examination of Toxoplasma cells where ERK7 was knocked down at protein level using an auxin-induced degron showed a catastrophic failure to assemble conoid. This phenotype is similar to the loss of MAPK15 (which is an ortholog of ERK7) in C. elegans where the loss of this kinase leads to catastrophic failure in the biogenesis of cilium (1). Similarly, ERK7 is involved in phosphorylation of an actin regulator to bring about the biogenesis of cilia in Xenopus embryos (2).
An unequivocal proof for the role of ERK7 in conoid biogenesis was found when wild type ERK7 was ectopically expressed in Toxoplasma cells lacking ERK7 protein and conoid biogenesis was found to be resumed (3). However, ectopic expression of a kinase-dead mutant of ERK7 could not rescue the phenotype. These findings suggest that obligate Apicomplexan parasites such as Toxoplasma originated from free-living protist ancestors and throughout evolution, the cellular machinery has undergone remarkable adaptations for a parasitic mode of life. A Cilium has undergone dramatic modifications to become the invasion machine known as Conoid but the mechanisms of its biogenesis and protein cascades involved in its assembly have remained conserved during its evolutionary journey to become a villain to manifest parasitism.
Suggested readings:
- Miyatake K, Kusakabe M, Takahashi C, Nishida E. ERK7 regulates ciliogenesis by phosphorylating the actin regulator CapZIP in cooperation with Dishevelled. Nat Commun. 2015; 6:6666. Published 2015 Mar 31. doi:10.1038/ncomms7666
- Kazatskaya A, Kuhns S, Lambacher NJ, et al. Primary Cilium Formation and Ciliary Protein Trafficking Is Regulated by the Atypical MAP Kinase MAPK15 in Caenorhabditis elegansand Human Cells. Genetics. 2017;207(4):1423-1440. doi:10.1534/genetics.117.300383
- O'Shaughnessy WJ, Hu X, Beraki T, McDougal M, Reese ML. Loss of a conserved MAPK causes catastrophic failure in assembly of a specialized cilium-like structure in Toxoplasma gondii. Mol Biol Cell. 2020;31(9):881-888. doi:10.1091/mbc.E19-11-0607






