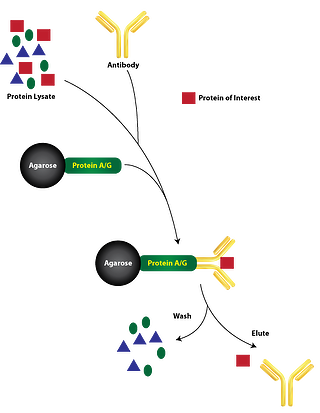
 The cardinal principle behind immunoprecipitation (IP) is the isolation of an antigen by interaction between the antigen and a specific antibody conjugated to a sedimentable matrix. Antigen source may vary from cells or tissues without labeling, radio labeled or tagged cells, labeled or unlabeled proteins isolated from subcellular fractionation or in-vitro translated proteins. Immunoprecipitation can also be helpful in protein fraction analysis separated by gel filtration or differential sedimentation based on density gradients. In order to capture interacting antigens, antibodies developed from various animal species that could be either polyclonal or monoclonal, are used for immunoprecipitation. These antibodies are tagged non-covalently with specific immunoadsorbents such as protein A–or protein G–agarose or covalently linked with a solid-phase matrix.
The cardinal principle behind immunoprecipitation (IP) is the isolation of an antigen by interaction between the antigen and a specific antibody conjugated to a sedimentable matrix. Antigen source may vary from cells or tissues without labeling, radio labeled or tagged cells, labeled or unlabeled proteins isolated from subcellular fractionation or in-vitro translated proteins. Immunoprecipitation can also be helpful in protein fraction analysis separated by gel filtration or differential sedimentation based on density gradients. In order to capture interacting antigens, antibodies developed from various animal species that could be either polyclonal or monoclonal, are used for immunoprecipitation. These antibodies are tagged non-covalently with specific immunoadsorbents such as protein A–or protein G–agarose or covalently linked with a solid-phase matrix.
The general protocol of Immunoprecipitation consists of following steps which can further be modified based on the antigens or matrix:
Stage 1 Antigen solubilisation: In order to solubilise antigens from the membrane or cells, non- denaturing detergent containing lysis buffers can be used. Cells grown in suspension culture or on tissue culture dishes can be lysed with non- denaturing detergent. Cells can also be lysed with denaturing lysis buffer alternatively. Mechanical disruption or sonication is used in for extracting soluble proteins from animal tissue without denaturing conditions. Although, disruption of cell wall is required to extract antigens from yeast cells.
Stage 2 Antibody coupling with matrix : In this step desired antibody is coupled either covalently or noncovalently to a sedimentable, solid-phase matrix in order to allow separation from the solution using low-speed centrifugation. Generally, antibodies are non-covalently attached with protein A–or protein G–agarose or covalently coupled with agarose.
Stage 3 Immunoprecipitation: The final step of the process is achieved by incubating the solubilized antigen with the immobilized antibody, subsequently washed extensively in order to remove unbound proteins. Alternatively, the immune complexes may be precipitated by using antibodies contained in an anti-immunoglobulin (anti-Ig) serum. Further the immunoprecipitated antigens could be detached from the antibodies and reprecipitated by a method called “immunoprecipitation-recapture”. This method has few advantages over the conventional IP. It can be used with the same antibody for subsequent purification of the antigen, or a second antibody can be utilized to study protein-protein interactions or to recognize components of multisubunit complexes.
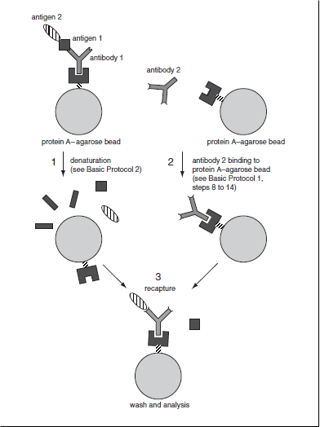 Immunoprecipitation-Recapture:
Immunoprecipitation-Recapture:
This method is bit advantageous over conventional IP methods as it allows identification of specific antigen in cases where the general IP gives ambiguous identification. Mechanistically, as the antigen is retrieved from conventional IP, the antigen can subsequently be dissociated from the beads and reimmunoprecipitated or recaptured using the same antibody or with a different antibody. This method allows analysis of subunit composition of multi-subunit or multi-protein complexes. The crucial step is the ability of the second step antibody to capture denatured antigens. In order to achieve dissociation of antigen from the beads, antigen-antibody-bead complexes are treated with SDS and DTT high temperature first. Subsequently, Triton-X containing solution is used to dilute SDS amount and excess iodoacetamide is utilized to neutralize DTT. Further recapture is done as conventional IP.






