Part 1: Essentials of protein phosphorylation
American chemist Westheimer asked an interesting question in 1987 in his very famous article published in ‘Science’ the same year, ‘why nature chose phosphates?’. He elegantly approached this problem from a chemists’ perspective. Phosphorus, a group 15 element, is an essential element of biomolecules. Phosphorus and its chemistry pervade cells and living systems, ATP, the ubiquitous currency of energy, is a nucleotide with two phosphates available in high energy phospho anhydride bonds. The backbone of genetic material (DNA or RNA) is made up of phosphates, two adjacent nucleotides are linked by phosphodiester bonds. The negative charge present on phosphodiester linkages in nucleic acids makes them resistant to hydrolysis and therefore very stable in aqueous environments, a key feature required for a molecule to qualify as a genetic material is chemical stability. Other group 15 elements like arsenic don’t make esters that are stable in aqueous conditions at neutral pH.
A large number of metabolites undergo phosphorylation to form corresponding phosphates in the cells and so do a large number of proteins. In this review we will quickly discuss the importance of phosphates and in particular, phosphorylation of proteins in cells.
One of the immediate and most obvious outcomes of phosphorylating a molecule is to impart negative charge to the molecule. Since phosphate occurs as a di-anionic moiety at cellular pH, charge imparted by phosphate group can bring about a change in conformation of the proteins by allosteric effects, it can also change the isoelectric point of proteins. In fact, phosphorylation can reduce net positive charge on a given molecule, which as we will explore further, can have interesting consequences for protein-lipid membrane or protein-protein interactions.
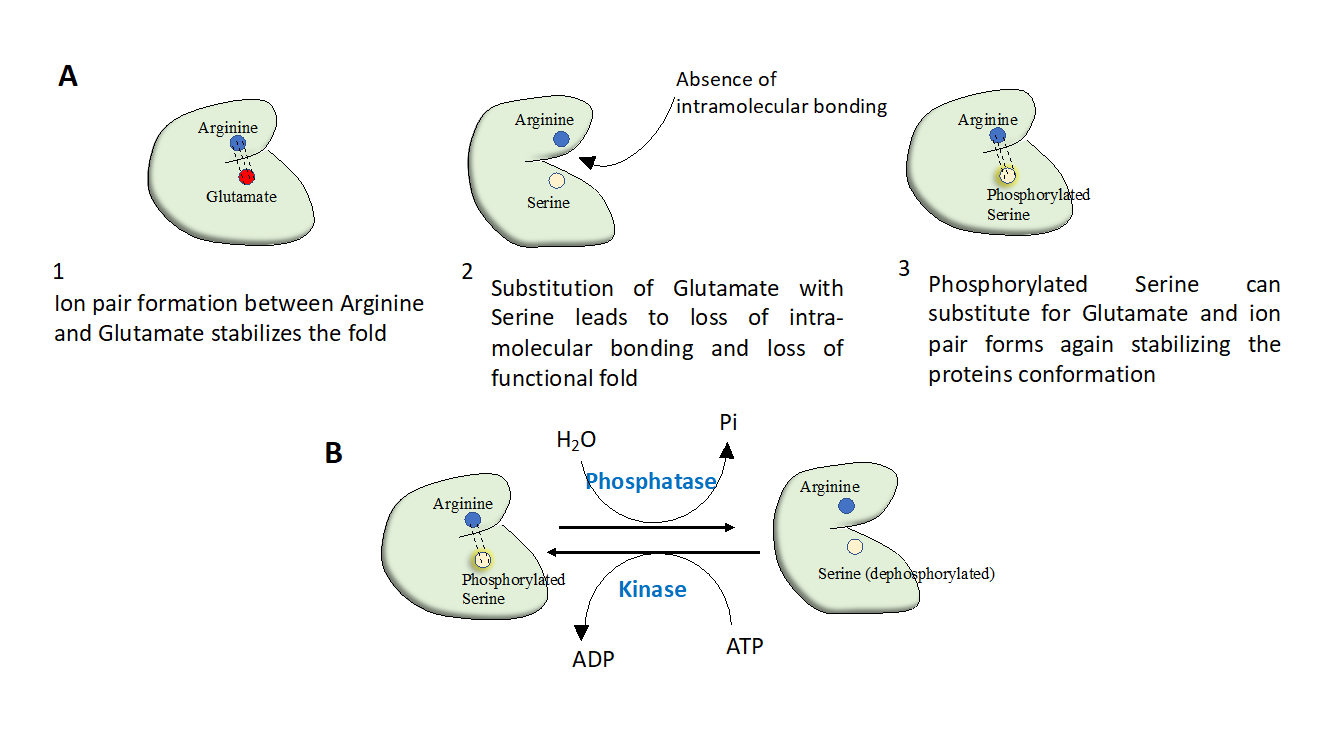
Figure 1: Protein phosphorylation evolved as a regulatory switch. A) During the course of evolution of proteins, negatively charged residues such as glutamate and aspartate have undergone mutations and got replaced by serine or threonine. This creates a contrivance to regulate protein structure by phosphorylation or de-phosphorylation of the serine/threonine residues. (B) Phosphorylation has emerged as a switch to regulate protein function; active state of protein can be switched on and off by only altering phosphorylation of a critical residue on protein. This is achieved in-vivo by concerted activities of kinases and phosphatases.
A large number of metabolites are rendered charged by phosphorylation and this occurs primarily to prevent diffusion of these molecules across plasma membrane. A charged molecule cannot leave plasma-membrane and remains inside the cell. Moreover, negative charge greatly diminishes the attack by nucleophiles.
Nine out of 20 standard amino acids can undergo phosphorylation but predominantly only serine, threonine, tyrosine (Hydroxylated amino acids) undergo phosphorylation in eukaryotes. The phosphorylated amino acids are very different from negatively charged amino acid residues, such as glutamate or aspartate. They have a larger solvation sphere and can form stronger ion pairs or salt bridges, even their hydrogen bonding potential is greater than these acidic residues. Protein phosphorylation has evolved to allow for a greater control of protein function, protein function can be switched on or off by altering charge on its surface by phosphorylation.
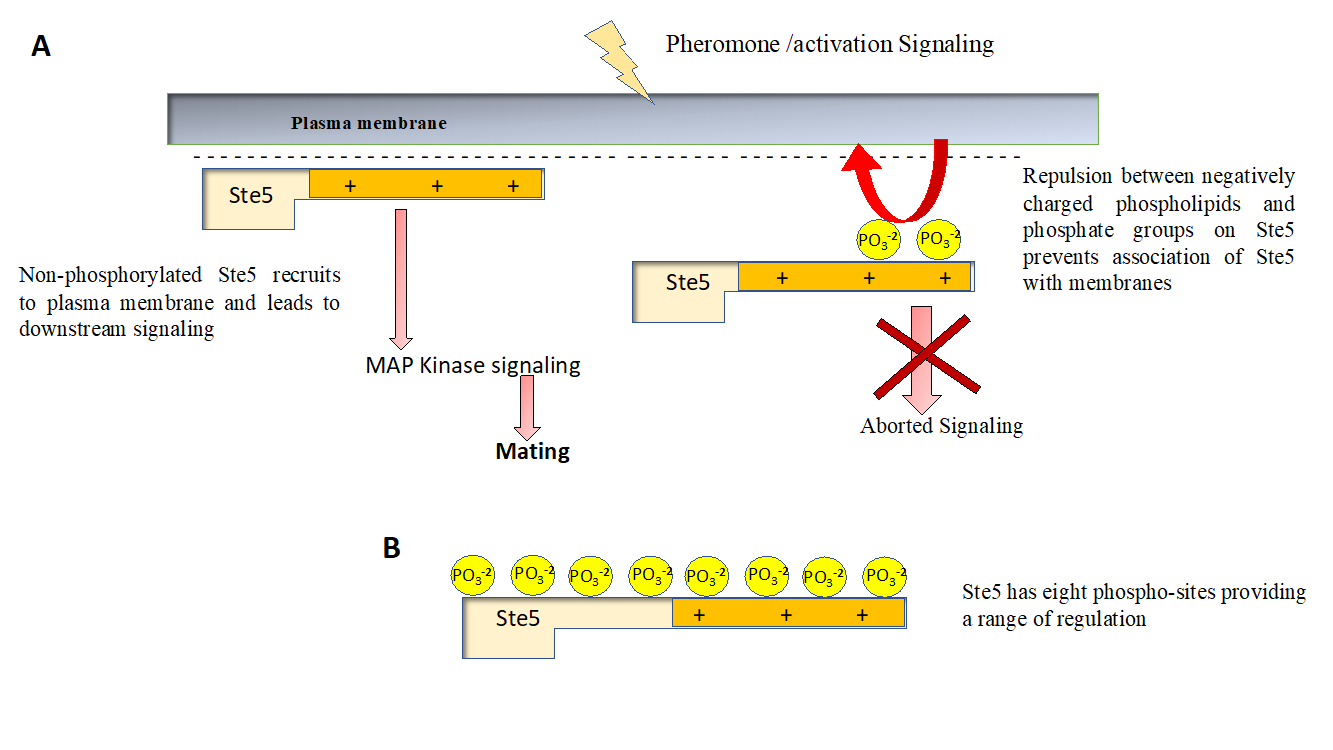
Figure 2 Regulation of signaling by protein phosphorylation (A) Ste5 is a well-studied signaling molecule that plays a critical role in relaying signal from pheromone to stimulate budding in yeast. Membrane recruitment of Ste5 is critical for its role as a signaling molecule, Phosphorylation of Ste5 prevents its membrane recruitment as it leads to a net negative charge on otherwise basic protein. Failure of Ste5 to recruit membrane impair it’s signaling function and mating doesn’t happen. (B) Ste5 has a basic motif (positively charged) that interacts with negatively charged phospholipids of the plasma membrane and also interacts with GPCR subunits (not shown for sake of simplicity). Ste5 has eight phosphorylation sites, allowing a graded response (and hence fine regulation) of its membrane residence time and hence signaling function, decreasing with each additional phosphorylation event.
The effect of phosphorylation on proteins vary dramatically, for example in case of Glycogen phosphorylase, the effect of phosphorylation is not additive (and also not inhibitory) unlike Ste5, but phosphorylation of a key serine residue in this enzyme leads to a dramatic change in protein conformation due to allosteric effects, leading to tetramerization of protein and activation of catalysis. Phosphorylation of certain proteins don’t cause any change in conformation but create sites for docking of other molecules such as binding of SH2 domain containing proteins on phosphorylated tails of cytokine receptors on cell surface or binding of 14-3-3 proteins to poly phosphoserine residues. Phosphorylation can influence protein function in many ways (highly context specific) and is by and large most pervasive post-translational modification and perhaps nature’s favorite way of decorating proteins.
Suggested readings/ References
- Westheimer FH. Why nature chose phosphates. Science. 1987 Mar 6;235(4793):1173-8
- Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2012 Sep 19;367(1602):2513-6.
- Serber Z, Ferrell JE Jr. Tuning bulk electrostatics to regulate protein function. Cell. 2007 Feb 9;128(3):441-4
- Good M, Tang G, Singleton J, Reményi A, Lim WA. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation.Cell. 2009 Mar 20;136(6):1085-97
- Pearlman SM, Serber Z, Ferrell JE Jr. A mechanism for the evolution of phosphorylation sites. Cell. 2011 Nov 11;147(4):934-46
- Nishi H, Shaytan A, Panchenko AR. Physicochemical mechanisms of protein regulation by phosphorylation. Front Genet. 2014;5:270. Published 2014 Aug 7.






