Proteins are phosphorylated and dephosphorylated all the times in the cells in a highly regulated manner. Protein phosphorylation and dephosphorylation are critical for signaling, cell division, protein translation, metabolism and survival. Activity of substrate proteins is tightly regulated by concerted activities of kinases and phosphatases in-vivo. On an average, roughly 3% of a typical eukaryotic genome encodes for kinases or phosphatases, yet more than 30% of the total number of proteins in a cell undergoes phosphorylation. As previously discussed, there is no general rule about effect of phosphorylation on proteins and same is true for dephosphorylation also. The effect of phosphorylation or dephosphorylation varies in context with different proteins.
What are Paradigms of regulation for kinases and phosphatases?
Cells maintain a basal kinase and phosphatase activity and chemical inhibition of either activity leads to dysregulation of protein function in cells, for example suppression of basal phosphatase activity in cells can amplify the basal phosphorylation events and can result in unwanted outcomes for the cells such as changes in cellular morphology or physiology or both, stress response activation and cell death and similarly inhibition of kinase activity would tip off the balance in the favor of abundance of dephosphorylated proteins. Apart from basal activity, activity of kinases and phosphatases are inducible in highly specific manner. The signaling pathways in cells are wired around kinases and phosphatases and therefore inducible activation of their activities is a major theme in signaling biology.
In some of the pathways, the cell surface receptor itself possesses intrinsic kinase or phosphatase activities in its cytoplasmic domain, whereas in many others the activated receptor interacts with kinase or phosphatase molecules upon stimulation. It is often perceived that kinases are always activator and phosphatases are always the desensitizers (inhibitory) in their mode of action, but that may not always be the case. For example, CD45 has an intrinsic tyrosine phosphatase activity, it is now established as an essential component of the signalling machinery of lymphocytes, it plays a critical role in antigen-stimulated proliferation of T lymphocytes and during the course of thymic development.
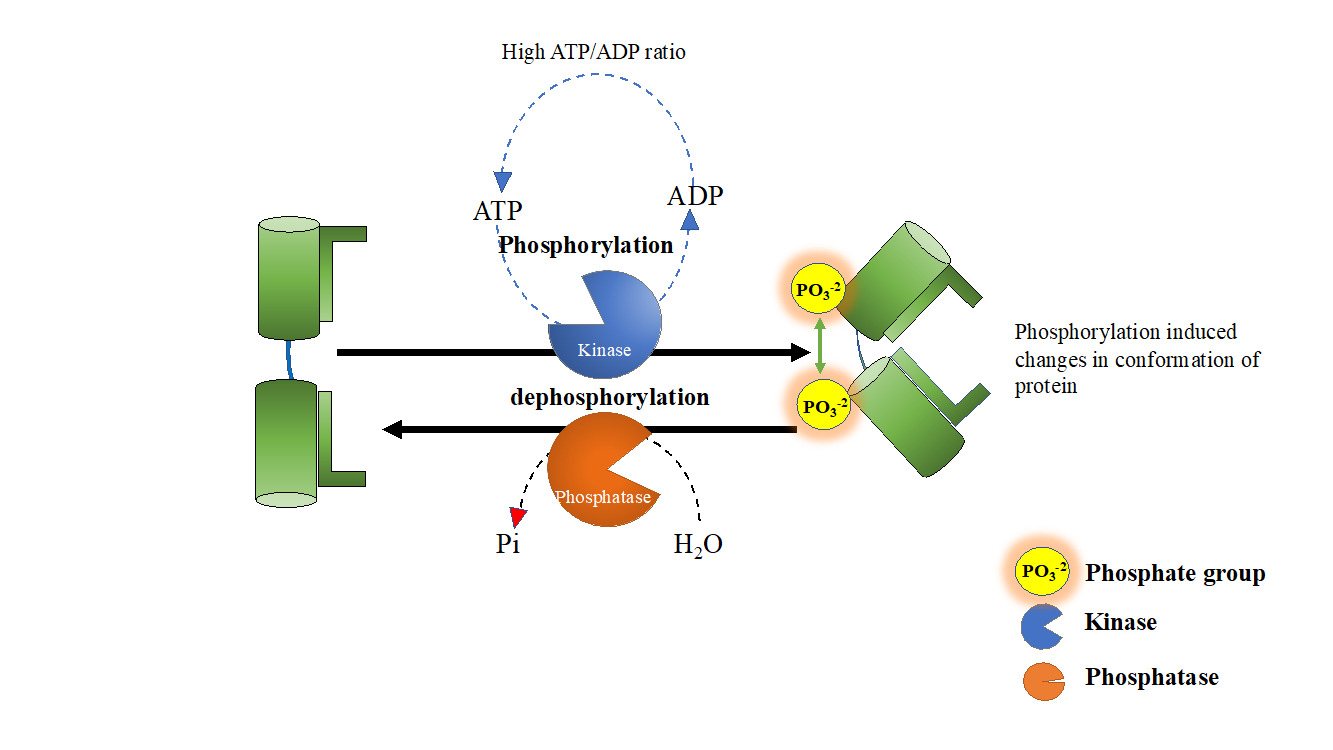
Figure 1. High ATP/ADP ratio in cells regulate in vivo kinase and phosphatase functions.
In fact protein kinases and phosphatases are both phosphotransferases, but in vivo their function is tightly regulated, phosphorylation is always catalysed by kinases whereas dephosphorylation is driven by phosphatases. Living cells maintain a high ATP/ADP ratio, and there is not enough ADP in cells to drive spontaneous dephosphorylation of phosphorylated proteins. Similarly, a high ATP concentration (2-4mM is typical ATP concentration in cells) prevents kinases from acting promiscuously as phosphatases. Therefore, dictated by high ATP/ADP cellular ratio, kinases and phosphatases are bound to behave as ‘bona-fide’ kinase and phosphatase. Protein phosphatases therefore drive forward phosphorylation reaction (See Figure 1) by replenishing dephosphorylated proteins. Presence of high ATP/ADP ratio in vivo also explain the altered behavior of kinases/phosphatases under in-vitro conditions.
Inside cells, kinases are wired into a hierarchy where kinases higher in hierarchy bring about activation of lower kinases and signal is thereby amplified at each step much like a cascade. Phosphatases usually don’t work that way, and therefore to maintain a balance, phosphatase activity is present in several fold excess of kinase activity, for example protein tyrosine phosphatase activities are in general two to four order magnitude higher than protein tyrosine kinase activities.
How do kinases maintain their specificity?
The inside of a cell is a highly crowded space with a myriad of molecules sloshing around. It is therefore critical for survival of cells that phosphorylation and dephosphorylation occurs at right place at the right time (See figure 2). Kinases exhibit exquisite specificity in terms of their substrate preferences. Serine, threonine and are phosphorylated by serine-threonine kinases and tyrosine residues are phosphorylated by tyrosine kinases. Kinases recognize their substrate proteins through two different mechanisms (1) proximal mode, where a kinase recognize a peptide sequence as a target, (2) Distal mode, a domain on substrate protein is recognized by a recognition/cognate domain on kinase molecules. Apart from these two modes of substrate identification, the kinases also ‘home-in’ on their substrate molecules by the help of adaptor proteins such as Beta-arrestins.
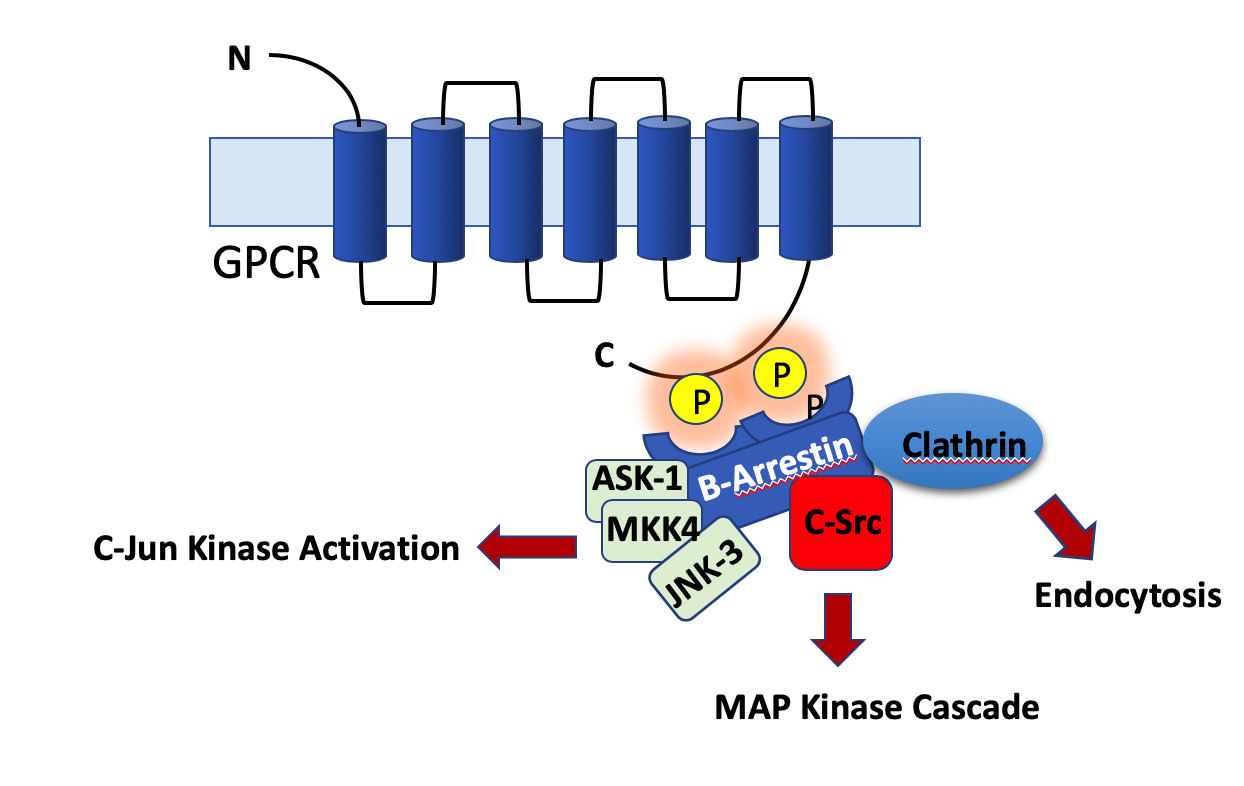
Figure 2. The Beta arrestins are adaptor proteins that bring about downstream signaling from GPCR proteins. They dock to phosphorylated cytoplasmic tail of GPCRs. The specificity of phosphorylation is critical since an aberrant phosphorylation can lead to aberrant activation of many signaling pathways.
Integration of responses
Kinases and phosphatases work together to ‘toggle’ a protein between phosphorylated and dephosphorylated states but as explained earlier that doesn’t imply that phosphorylation will activate signaling and dephosphorylation will inactivate signaling, Infact kinases and phosphatases can integrate together to positively or negatively regulate a signaling pathway. The effect of kinase and phosphatase activity on a substrate protein is a binary response but signaling pathways are not binary, how do then binary switches combine to give a range of signaling outputs in terms of phosphorylation of proteins. Any given kinase/ phosphatase population in cells can integrate together to give rise to a variety of responses or output in response to a given stimulus or input. Following are some commonly encountered paradigms,
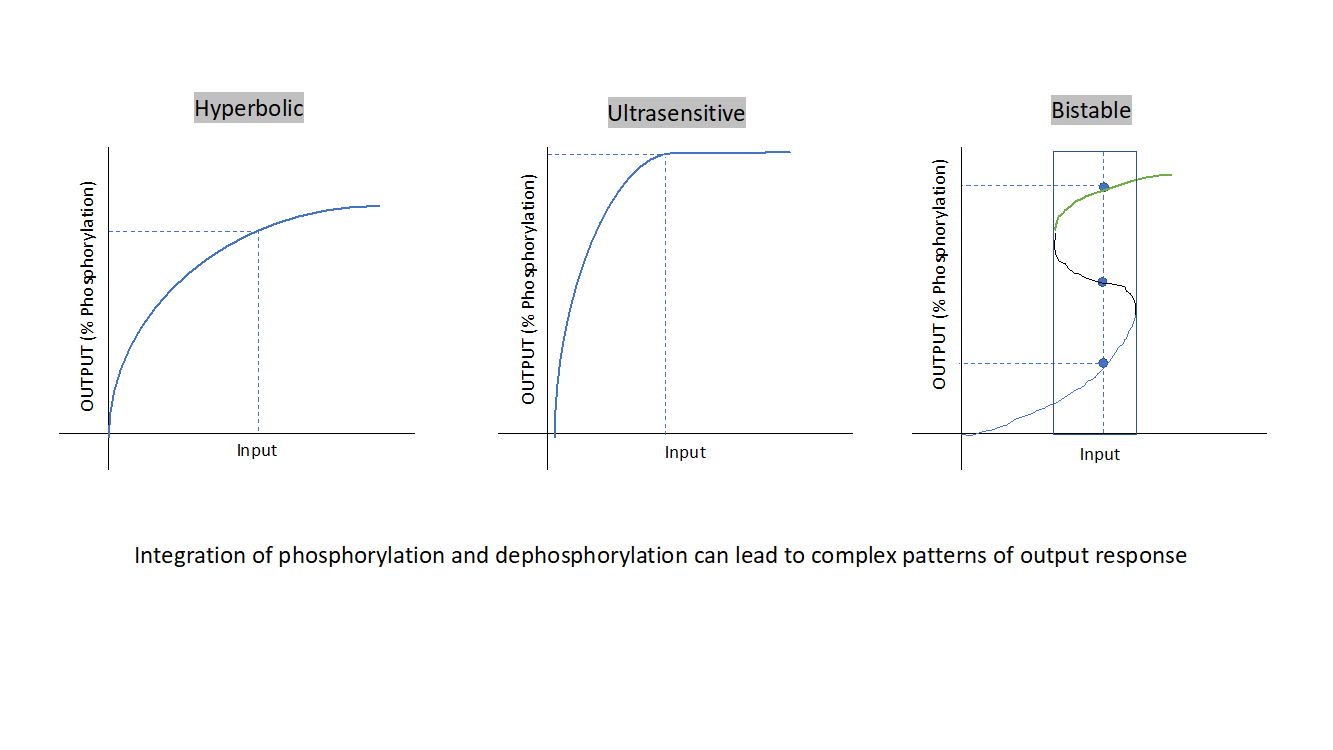
Figure 3: The integration of kinase and phosphatase activity in cells can lead to a variety of signaling responses as described above (from left to right), Hyperbolic response, Ultrasensitive response, Bi-stable response.
Hyperbolic response signaling, a hyperbolic response exists between input and phosphorylation of substrate proteins which can be achieved alternatively if the input decreases the phosphatase activity in a hyperbolic manner following mass action kinetics. In such a scenario changing kinase or phosphatase activity will have same effect on signal output. Hyperbolic response signaling allows modulation of response. But in cells, complex signaling networks exist to respond to a range of varied signal strengths, hyperbolic response signaling can’t generate a maximal response to a wider range of input values.
And there comes into picture another kind of signaling response, this is ultrasensitive signaling response. Here the input leads to activation of kinases and at the same time indirect suppression of phosphatase activity, known as reciprocal signaling, this leads to ultrasensitive response. As one could infer from the nature of curve, the response is flat at lower and higher input strength but is extremely sensitive to small change in inputs within a range in-between and a maximum response is generated. A maximal phosphorylation response is generated for a range of input values. Such signaling modules regulates mitotic entry of cells. Kinases and phosphatases can interact in many other ways other than reciprocal signaling to generate ultrasensitive response modules. This response can be approximated by Hill’s equation.
Cells have yet another mode of generating a signaling response by ingenious integration of kinase and phosphatase activity known as bi-stable response. This is usually achieved by employing feedback loops, for example auto phosphorylation of kinase further boosts its activity and similarly if it decreases its own dephosphorylation in response to its activation. The outcome is a bi stable response where a wide range of input values have a robust output response to small changes in input, anything below threshold leads to a stable low phosphorylated state and anything that leads to crossing the threshold will have a stable maximum phosphorylated state and hence the bi-stable nature. The signal cannot be modulated for intermediate levels of phosphorylation response. Such circuitry is found at important decision-making points such as mitotic checkpoints.
Cells can constrain kinases and phosphatases to have their activity in a given subcellular compartment and at a given time point by controlling their spatial and temporal localizations in the cells. This is achieved by a variety of diverse mechanisms which is in itself deserve a separate blog article and would be discussed elsewhere in future.
References
- Pawson T, Scott JD. Protein phosphorylation in signaling--50 years and counting. Trends Biochem Sci. 2005 Jun;30(6):286-90.
- Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995 Jan 27;80(2):225-36.
- Cheng HC, Qi RZ, Paudel H, Zhu HJ. Regulation and function of protein kinases and phosphatases. Enzyme Res. 2011;2011:794089.4.
- Lin J, Xie Z, Zhu H, Qian J. Understanding protein phosphorylation on a systems level. Brief Funct Genomics. 2010 Jan;9(1):32-42.5.
- Miller CJ, Turk BE. Homing in: Mechanisms of Substrate Targeting by Protein Kinases. Trends Biochem Sci. 2018 May;43(5):380-394.
- Good, M.C. et al. (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332, 680–686
- Gelens L, Qian J, Bollen M, Saurin AT. The Importance of Kinase-Phosphatase Integration: Lessons from Mitosis. Trends Cell Biol. 2018 Jan;28(1):6-21






