Biochemistry and Cell-biology owe a lot of their existence to the Subcellular fractionation techniques. Subcellular fractionation refers to disintegrating intact cells into their integral parts, the cellular organelles. For cell biologists, the key objective is to isolate each cellular organelle to a high degree of purity even if the quantify is small. This is necessary for undertaking electron microscopic studies to reveal ultrastructure of organelles. On the other hand, biochemists are more interested in macromolecular composition (such as proteins, lipids and carbohydrates) and hallmark enzymes of each subcellular fraction and therefore they seek pure fractions in sufficient quantity. The technology of the subcellular fractionation was pioneered by the Christian De Duve, Camillo Golgi and George Palade with significant contributions made by Albert Claude.
Based on requirements one has to choose an appropriate method and adapt it to the specific requirements of the experiment. Broadly, there are two methods to perform subcellular fractionation using centrifugation. They are 1. Differential centrifugation 2. Equilibrium density centrifugation
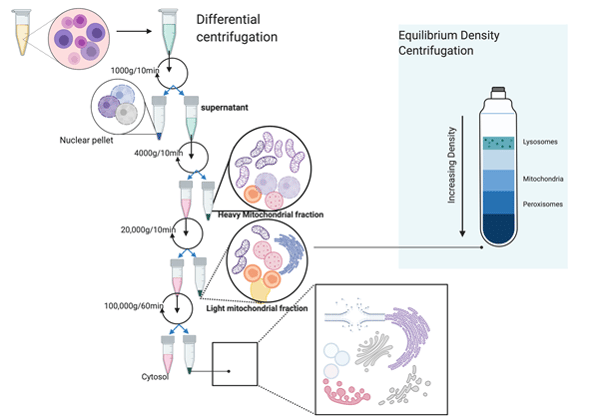
Figure 1. Differential centrifugation
Subcellular compartments/organelles vary in physical parameters such as mass, size and density. Differential centrifugation exploits the sedimentation behaviour of particles varying in their densities. The particles are separated based on their differential sedimentation velocities in a medium of considerably small density (ρ2) and low viscosity. The density of the medium is critical because a high-density medium will prevent sedimentation of cellular organelles and the organelles will float as predicted by the equation given below.
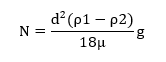
ν =Sedimentation velocity
d=diameter of distance
ρ1=density of particle
ρ2=density of medium
g=centrifugal force
µ=Viscosity of medium
Often sucrose (0.25M) is used as a medium, this is because sucrose has the ‘right’ density to bring about sedimentation of cellular organelles. Moreover, sucrose at 0.25 M doesn’t have a high osmolarity and viscosity. The consideration of osmolarity is important because hyperosmotic concentrations of the medium can lead to osmotic-shrinkage of organelles. The presence of ions and detergents can also change the sedimentation behaviour of subcellular organelles. The differential centrifugation leads to separation of cellular lysate into a pellet and a supernatant fraction. At each step, the pellet fractions are made up of heavier fractions that settle and make a pellet at a given ‘g-force’ and time interval whereas supernatant is made of parts that could not be pelleted at that g-force and time values.
Another important factor is the volume of the sample to be fractionated. The resolution achieved in differential centrifugation is volume dependent. Particles at the top of the tube have to travel maximum distance to make a pellet at the bottom of the tube and are exposed to lower values of g force. However, the heavier particles near the bottom of the tube are exposed to higher values of ‘g-force’ and have the shortest distance to travel. This leads to low yield and also contamination for lighter or smaller particles. The figure shows the scheme of differential centrifugation to obtain different subcellular organelles.
2. Equilibrium density centrifugation
Equilibrium density centrifugation also known as gradient centrifugation is often employed to separate cellular components that have closely related densities. A non-ionic medium of low density is chosen which also has a low osmolarity and viscosity for similar reasons explained earlier. Usually, Sucrose and Glycerol have been medium of choice but lately synthetic mediums such as Nycodenz, Iodixanol and Percol have been also used with great success and better resolving ability. Sucrose and glycerol are limited by the fact that if their concentrations are increased (to adjust the medium density, ρ2 see equation above) it leads to a quick jump in osmolarity which is not suitable for obtaining intact organelles. Even at high concentrations Percol, Nycodenz and Iodixanol don’t have a sharp change in osmolarity.
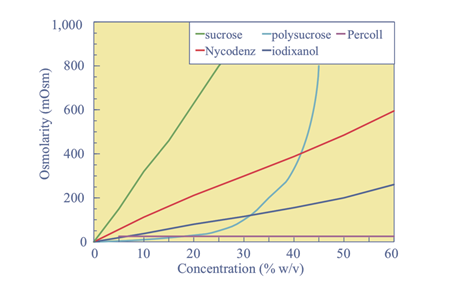
Figure 2. Adapted from: Fractionation of Subcellular Organelles
John M. Graham, Current Protocols Current Protocols in Cell Biology 3.1.1-3.1.22, December 2015
For equilibrium density centrifugation, the samples are spun at high ‘g-force’ values (40,000-50,000) for almost 60 minutes, this generate zones of varying densities inside the tube and the organelles or subcellular fractions are separated on basis of their densities in the gradient medium, each organelle in a zone equal to its density.
G-biosciences offers a kit for subcellular fractionation applications known as FocusTM SubCell which can be used for subcellular fractionation with a range of biological samples.





