In eukaryotic cells, RNA processing occurs in the nucleus and the functional mRNA is transported to the cytoplasm. As soon as the mRNA transcript is synthesized from RNA polymerase II, RNA binding proteins (RBPs) attach to the mRNA transcript to prevent it from degradation during processing and exporting it from the nucleus. RBPs play an important role in RNA stability, maturation, transport, and localization. This process results in the formation of stable mature mRNA (lasts for several hours) than that in prokaryotes (5 seconds).
The three major steps in eukaryotic RNA processing are:
- 5’ Capping,
- 3’ Cleavage / Polyadenylationand
- Pre mRNA Splicing
In this process, a 7-methylguanosine cap is attached at the 5’end by a phosphate linkage followed by the addition of a poly-A tail and finally removing introns from the pre mRNA resulting in mature functional mRNA.
- 5’ Capping:
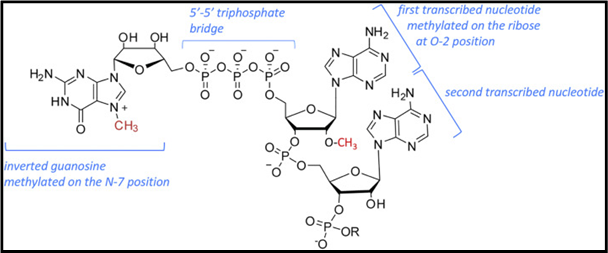
During the initiation of mRNA from RNA polymerase II, a 7-methylguanosine cap is attached at the 5’ end of the growing transcript. This process is catalyzed by a dimeric capping enzyme. The 5’ cap prevents the nascent mRNA from degrading and also facilitates the initiation of the ribosomal translation by protein synthesis factors. The structure of 7-Methylguanosine is similar to the guanine nucleotide, but with an additional methyl group.
Fig 1: 5’ mRNA Cap. (Source: Alison et al, BBA, 2019)
3’ Cleavage / Polyadenylation:
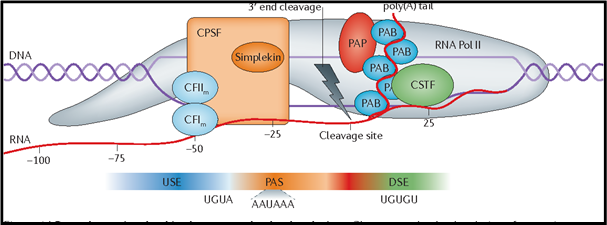
Polyadenylation is the process in which 200-250 Adenine residues are attached to the 3’ end of pre mRNA transcript. Except for histone mRNAs, all other mRNAs are polyadenylated. All mRNAs consist of two conserved sequences necessary for cleavage and polyadenylation located near 3’ cleavage site where the poly-A tail is formed.
- AAUAAA sequence, 20-30 nucleotides upstream.
- GU rich or U rich sequence, 50 nucleotides downstream.
Cleavage and polyadenylation specificity factor (CPSF) binds to the upstream AAUAAA sequence followed by binding of Cleavage stimulatory factor (CStF), Cleavage Factor I (CFI) and Cleavage Factor II (CFII). CStF interacts with downstream GU or U rich sequence and stabilizes the multi-protein complex. Finally, before the cleavage starts, poly (A) polymerase (PAP) binds to this complex to facilitate polyadenylation as soon as the cleavage starts.
Polyadenylation takes place in two phases:
- A slow phase where around 12 Adenine residues are added byPAP and,
- A rapid phase where 200-250 Adenine residues are added byPAP stimulated by PAB II.
Multiple copies of Poly(A)-binding protein (PABII) binds to the short polyadenine tail of mRNA and stimulates PAP to accelerate the polyadenylation. And also signals PAP for the termination of polyadenylation.
Fig 2: 3’ Cleavage of pre-mRNA (Elkon, Ran et al. Nature Reviews Genetics, 2013)
- Pre-mRNA Splicing:
The final step in RNA processing is “Splicing”, where Introns (non-coding intervening sequences) are removed and Exons (coding sequences) are spliced together to form mature, functional mRNA. The resulting mRNA contains only Exons. Initial studies showed that these introns are present in viral and eukaryotic genome but not in the prokaryotic genome.
The sequences that are present in eukaryotic genomic DNA and absent in cDNA are Introns. Intron sequences are flanked by conserved sequences 5’ GU and 3’ AG dinucleotides. Introns are removed from the pre mRNA transcripts by cleavage at these conserved sequences called ‘Splice sites’. Upstream from the 3’ end of the Intron, another conserved pyrimidine rich sequence ‘YNYYRAY’ is present, called Branch point. (Where Y=Pyrimidine, N=Nucleotide, R=Purine, and A=Adenine.)
Exon splicing is mediated by two sequential trans-esterification reactions in which one phosphate-ester bond is exchanged. Five U-rich Small nuclear RNAs (snRNAs-U1, U2, U4, U5, U6) assist in mRNA splicing. Splicing occurs in a cellular machine called small nuclear ribonucleoprotein particles (snRNPs), in the nucleus. snRNP U1 attaches to the intron as its 5’ end is complementary to the 5’ end of Intron resulting in the cleavage of pre mRNA at 5’ end of Intron. The cut end attaches to the branch point (G of splice site to A of Branch point) and forms “Lariant”, a looped structure.
The snRNPs U2 and U4/U6 help in the positioning of 5’end and branch point in proximity and snRNP U5 help in cutting, joining of 3’end to 5’end, and positioning in proximity. Thereby the neighbouring exons are covalently bound and resulting lariant with U2, U5, and U6 is released.
In addition to these sequences, eukaryotic genes (on Exons) contain Exonic Splicing Enhancers (ESEs). ESEs help position the splicing apparatus and bind proteins involved in recruiting splicing machinery to the correct site.
In general, splicing occurs between exons on a single RNA transcript, but in some cases, exons on different pre mRNA transcripts ligate together as “Trans Splicing.”
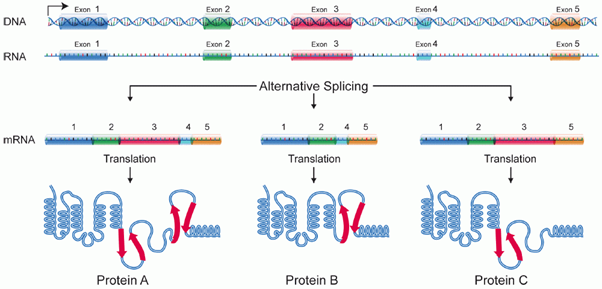
“Alternate Splicing” as the name suggests, one pre-mRNA molecule could be spliced at different sites resulting in a variety of mature mRNAs with different exon combinations. This alternate splicing was first discovered in Adenovirus in 1977 and later observed in higher eukaryotes. Wang et al suggested that more than 90% of human genes are alternatively spliced. For instance, the same pre-mRNA in the diagram below can be spliced in three ways, depending on the retained exons. As a result, three different mature mRNAs are produced, every one of these mRNAs can be translated into a differently structured protein.
Fig 3: Alternative Splicing (Source:National Institute of Health)
The existence of Introns and alternate splicing helps in explaining the evolution of new genes. New exons can be inserted into old introns creating new proteins without disrupting the function of the old gene. Maybe this is one of the reasons how SARS-CoV-2 is different from the other coronaviruses.
Reference:
- Lodish H, Berk A, Zipursky SL, et al. Molecular Cell Biology. 4th edition. New York: W. H. Freeman; 2000. Section 11.2, Processing of Eukaryotic mRNA.Available from: https://www.ncbi.nlm.nih.gov/books/NBK21563/
- Alison Galloway, Victoria H. Cowling, mRNA cap regulation in mammalian cell function and fate, Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, Volume 1862, Issue 3, 2019, 270-279.
- Elkon, Ran et al. “Alternative cleavage and polyadenylation: extent, regulation and function.” Nature Reviews Genetics14 (2013): 496-506.






