 A restriction enzyme (restriction endonuclease) is a special enzyme that recognizes a specific sequence of nucleotides and cleaves DNA at that specific site (restriction site or target sequence). These enzymes, which are usually found in bacteria and other prokaryotes, are considered as one of the most important tools in recombinant DNA technology since they can easily cut DNA into fragments and/or join DNA molecules from different genomes so researchers can identify and characterize genes and examine gene expression and regulation.
A restriction enzyme (restriction endonuclease) is a special enzyme that recognizes a specific sequence of nucleotides and cleaves DNA at that specific site (restriction site or target sequence). These enzymes, which are usually found in bacteria and other prokaryotes, are considered as one of the most important tools in recombinant DNA technology since they can easily cut DNA into fragments and/or join DNA molecules from different genomes so researchers can identify and characterize genes and examine gene expression and regulation.
Cutting and Pasting with Restriction Enzymes: How Does It Work?
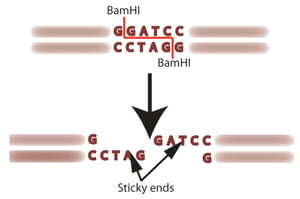
When a restriction enzyme finds its restriction site or target sequence (usually a sequence of 4 to 8 nucleotides), it will stop and cleave the DNA molecule (i.e., enzyme digestion). Since the recognition sequence is usually found on both strands (but running in opposite directions), the enzyme will cut both strands.
Some restriction enzymes produce blunt ends (the target sequence is cut right in the middle, leaving no overhangs) while others produce sticky ends (DNA fragments with one-stranded overhangs). Fragments with sticky ends are more useful in recombinant DNA technology since they can easily be joined together with ligase, cultivated in bacterial cells and used in a number of applications.
7 Things to Consider Before Making the Cut
If you’re not careful enough, restriction enzymes can behave erratically in the lab. Here are some factors you may need to consider when using them.
- Most restriction enzymes perform optimally at 37oC. Enzymes derived from microorganisms that thrive in extreme conditions show maximum activity at higher temperatures.
- Most enzymes perform best between pH 7.2 and pH 8.5. Star activity may increase outside the optimal pH range.
- BSA is commonly used in stabilizing restriction enzymes during digestion reactions since it protects the enzyme from non-specific adsorption, the adverse effects of proteases, and other damaging factors (e.g., intense heat, interfering substances, surface tension).
- Some enzymes are unable to bind with methylated recognition sites. This can be a problem since specific CpG islands are often methylated when a bacteria replicates a plasmid. To avoid running into this problem, choose a methylation-incompetent E. coli strain when propagating your plasmid.
- Star Activity. Long incubation periods and less than optimal buffer conditions (e.g., high glycerol concentration, low magnesium concentration, incorrect pH) can increase the chances of producing unexpected results. This phenomenon, known as star activity, may cause the enzyme to cleave the DNA at incorrect sites and/or facilitate single base substitutions.
- Base Pairing. To ensure efficiency, make sure your enzymes have at least six base pairs on either side of the recognition site. This is especially important when you are digesting the ends of a PCR product and/or when doing a double digest.
- Isoschizomers and Neoschizomers. There are times when you need to use restriction enzymes that recognize the same target sequence but exhibit a totally different property to get the results you want without increasing the risk of star activity and/or methylation sensitivity. This is when isoschizomers and neoschizomers come in quite handy. By definition, isoschizomers are different restriction enzymes that recognize the same DNA sequence and cleave at the same site while neoschizomers are enzymes that recognize the same sequence but cleaves at different positions.
Working with Restriction Enzymes? Consider These Tips
- Always wear gloves to avoid contaminating your samples. It can also help ensure your safety.
- Know your enzymes inside out. Knowing what makes it work can help you get the results you need and help you avoid problems along the way. For best results, check with the manufacturer for specific guidelines on using your chosen enzyme.
- Avoid overloading your digest reaction when running your gel. It will make it hard for you to distinguish the digested bands.
- Glycerol is often added to enzyme storage buffers to prevent freezing, but since high glycerol concentration interferes with enzyme activity, you need to dilute it to less than 5% before using it.






