In all the complex biological processes, the mechanism underlies a synchronized and orchestrated molecular organization, which works through protein-protein interactions. These interactions could be transient, such as catalytic or signal-transduction pathways, requiring a transitory protein-protein association. Alternatively, stable or semi-stable multi-protein complexes are also formed in many biological functions. Hence, in order to identify transient and semi-stable protein associations, chemical cross-linking of proteins came as a useful technique. It involves the formation of chemical covalent bonds between the interacting protein utilizing bifunctional reagents consisting of reactive end groups, which can react with the functional groups of amino acid residues, such as primary amines and sulfhydryls. The formation of cross-links provides direct and concrete information regarding the identity of the interacting proteins as well as the regions of contact between the proteins.
Currently, many assay and purification methods have been developed to study protein interactions and applications. One of the most innovative and useful way is to immobilize proteins onto glass surfaces derived from aminosilane reagent containing primary amine (-NH2). Glass substrates are thermally and mechanically stable, hence can be utilized as a microarray platform for DNA as well as protein biomarker detection.
The free amine groups of the glass surfaces are reacted with the heterobifunctional cross-linkers such as Sulfo-SMCC, which eventually results into an activated glass surface. These activated cross-linked surfaces can be used to bind with the sulfhydryl groups of the interacting proteins or antibodies.
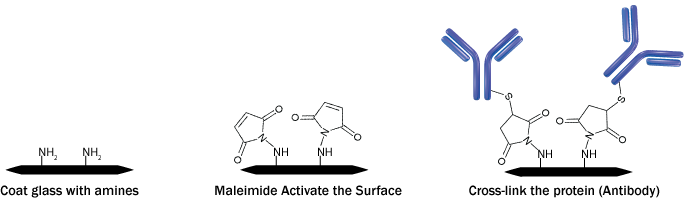
Figure 1: Protein cross-linking on a glass surface
Step1: Aminosylation of the glass surface
In order to make an activated glass surface, 2% aminosilane reagent is used in acetone. Cleaned glass, silica or quartz surfaces can be used for coating. The surface to be coated should be immersed with the coating agent for few seconds to few minutes, subsequently washed with acetone and dried in air.
Step 2: Modification of silylated surface
Aminosylated glass surface can further be processed with the addition of functional groups to attach proteins by either amine or sulfhydryl group attachment. Both of the methods differ with the sequence of addition of cross-linking functional group.
- Amine-to-Surface Attachment of the Protein
- Modification of the protein and silylated glass surface with the cross-linker
Purified protein is dissolved in a coupling buffer, such as PBS. Sulfo-LC-SPDP cross-linker solution is preferred to be prepared separately and fresh in water. Sulfo-LC-SPDP is a long chain, heterobifunctional, thiol-cleavable and membrane impermeable cross-linker. It consists of an amine-reactive NHS ester, which can react with the lysine residues and form a stable amide linkage. The opposite end of the space arm consists of a pyridyl disulfide group which will react with the free sulfhydryls groups resulting into a detectable by-product at 343 nm. Since the cross-linker is impermeable to the membrane, it allows cell surface labelling as well. The spacer arm is easily cleavable with mild denaturing treatment. Other heterobifunctional cross-linkers that react with amines and sulfhydryls can also be used, selected ones with long spacer arms and, if required, are cleavable.
Once the cross-linker solution is ready, it is mixed with the protein solution and incubated at room temperature for 30 minutes approximately. Similarly the silylated glass surface is incubated with Sulfo-LC-SPDP solution in the coupling buffer to attach the crosslinker on its surface. - Purification of the modified protein
The by-products of the previous step can be removed from the modified protein by dialysis or gel filtration. Similarly, the modified and activated glass surface can be washed with the coupling buffer to remove all the by-products. The modified surface is subsequently reduced using DTT for 30 minutes at Room temperature. The reduced surface should be further washed with coupling buffer to remove residual DTT. - Protein cross-linking on the reduced surface
In order to cross-link the modified protein with the reduced glass surface, the glass surface is incubated with the modified protein solution of approximately 18 hours at RT or 4°C. Furthermore, the surface is washed with coupling buffer in order to remove non-conjugated protein.
- Modification of the protein and silylated glass surface with the cross-linker
- Sulfhydryl-to-Surface Attachment of the Protein
- Modification of the silylated surface with Sulfo-LC-SPDP
The silylated glass surface is incubated first with the Sulfo-LC-SPDP solution (prepared in coupling solution) for approximately 30 minutes at room temperature. The SPDP-modified surface should be washed thoroughly with the coupling buffer to remove by-products and unconjugated cross-linker. - Availability of free sulfhydryls on protein
In order to assess the availability of free sulfhydryls on the purified protein, Ellman's reagent can be used . If the availability of free sulfhydryls is low, the protein solution can be reduced by incubating with 50 mM DTT for 30 minutes. Subsequently, reduced protein is purified by gel filtration.
- Protein cross-linking on the SPDP modified surface
The SPDP-modified surface is allowed to react with sulfhydryl-containing protein solution for 18 hours at room temperature or 4°C. The unbound protein and cross-linkers should be removed by washing in the coupling solution.
- Modification of the silylated surface with Sulfo-LC-SPDP
Step 3: Cleavage of Cross-linked protein from the surface after use
The surface bound proteins can be cleaved from the cross-linker by either using 10-50 mM DTT at pH 8.5 for 15-30 minutes at 37°C or boiling the protein-containing surface for 5 minutes in SDS-PAGE sample loading buffer.
Do`s and Don'ts:
- Solutions (Coupling solution and cross-linker solution) must be prepared fresh before doing the procedure.
- If not used immediately, the air-dried silylated surface can be stored for further use.
- The longer incubation of cross-linker solution with either the protein or silylated surface does not adversely affect the efficiency of the procedure. Though optimization of the incubation time is essential.
- The reduced surfaces can be dessicated and stored at 4°C if not used immediately.
- After every incubation the surface should be rinsed thoroughly with the coupling buffer in order to remove unconjugated products.
- Conjugation efficiency also depends on the availability of protein present for conjugation. It is always advised to take ample amount of protein and check its concentration before conjugation. In order to determine the efficiency, the washed out non-conjugated protein fraction can be assayed for concentration and compared to the pre-coupled fraction.






