There has been an exponential interest developing from past few years for studying the translational regulation of different genes in many model systems. Protein synthesis is the functional and fundamental biological process essential for decoding the genetic information and culminates it with a specific phenotype. Since the energy expenditure on protein synthesis is roughly more than half of the total energy expense of the cell during rapid growth, thus studying the kinetics of translational instrumentation has gained a broad attraction recently. As it has been shown previously, even a subtle altercation in translational apparatus can substantially affect human health. Hence, it is essential to profile the actively translating proteins comprehensively in order to develop insights into the cumulative translatome and its functional correlations.
Ribosome footprint profiling has come up as novel tool that can measure ribosomal abundance and the translation at a genome-wide level, thus addressing the global expression and its culmination into translational regulation along with the mRNA abundance. This method is very precise in delineating the translating regions of the genome providing the full coding potential of the cell.
The method is principally based on deep sequencing of the mRNA fragments protected by strings of ribosomes. Consequently it provides an idea of all the ribosomes that are active in a cell in response to a stimulus at specific time point. Collaterally, it also gives insight about what all proteins are actively getting translated at that point. It is a very systematic method of monitoring translation process and its correlation of protein abundance at the experimental time point.
While translating, each ribosome covers a small 29 nucleotide long fragment of the mRNA, hence protecting that fragment from digestion with exogenous RNAse. Therefore, if the sequence of the transcriptome is known, then the location of ribosomes in the translating fragment by using RNAse digestion can be determined, followed by isolation of the ribosome-protected fragments and determination of the sequence of protected fragment. By knowing the quantal distribution of the ribosome footprints on a known mRNA fragment, it can be easier to decipher the ribosomal movement and the degree of mRNA translation.
In general, ribosome footprinting consists of fabrication of two different RNA-seq libraries:
- mRNA fraction library to measure mRNA abundance
- Ribofraction for snapshot of the part of the transcriptome which is in the phase of active translation by ribosomes.
Conventional protocol for ribosomal footprinting has the following steps:
- Cell Lysate Preparation: Ribosomal footprinting can be used for adherent cell lines and tissue samples. An optional way is to add translational inhibitors before making cell lysate, although it depends on the type of the experiment and optimization of the protocol. It is an advantageous step as translation initiation inhibition cause accumulation of ribosomal footprints close to the start codon allowing the genome-wide identification of the translation starting site. Ideally, the lysis buffer should be 4 °C and lysis should be performed in the dish in order to avoid loss of sample and protein degradation. The cell lysate should be centrifuged before further procedure to remove cell debris and membrane fraction of the cell.
- Nuclease footprinting and ribosome recovery: In this step, digestion with RNaseI has been optimized by the experimenter as different commercial suppliers use different unit definitions to measure the activity of RNaseI. The activity variation of RNaseI can considerably alter footprinting output for a particular experiment. Consequently, it is advised to match the RNA-to-RNase ratio controls for an efficient footprinting. Important point to note here is that, some portion of undigested cell lysate has to be stored for control RNA-seq library which eventually can be used for calculating translational efficiency. Ribosomal and RNA fraction can be separated from the digested samples by using Sucrose-gradient based Ultracentrifugation in a TLA100.3 rotor at 100,000 rpm, 4°C for 1 h or 70,000 rpm for 2 h. RNA can be purified from the ribosomal pellet by using commercial kits.
- Footprint fragment purification: The crucial step here to follow is maintaining RNase free conditions during electrophoresis. Therefore, all the preparations must be done under RNase decontaminated conditions. The gel has to be prepared keeping in mind the size of footprints which might range from 20 nucleotides to larger fractions. In order to capture both small and large fractions, the fragments must be separated on a purification gel or column chromatography. The total RNA present in fragments is then eluted using RNA elution methods. In order to concentrate the samples, it can be precipitated further.
- Footprint fragment processing: The eluted fragments are processed further for making libraries. Footprint fragments are dephosphorylated and circularised using T4 PN Kinase. Dephosphorylation prepares the fragments for ligation to a linker DNA. The critical part here to follow is to polyadenylate the linker with a unique sample barcode for each pooling sample. The ligated fragments should be purified using concentration columns, although in order to minimise the loss of sample and multiplex sequencing, the ligations with different barcodes can be pooled together.
Ligated footprint fragments have to be converted into cDNA using Reverse Transcriptase followed by circularization for making pooled or unique cDNA footprint libraries. Circularized cDNA furthermore quantitated using qPCR in order to get a sense of optimal conditions for single library construction PCR
After single library synthesis it is crucial to analyse the length distribution, quality and concentration of the constructed library for further processing. After this, the constructed libraries can be sent for high-throughput sequencing. It is vital to use the sequencing paradigm with greater depth.
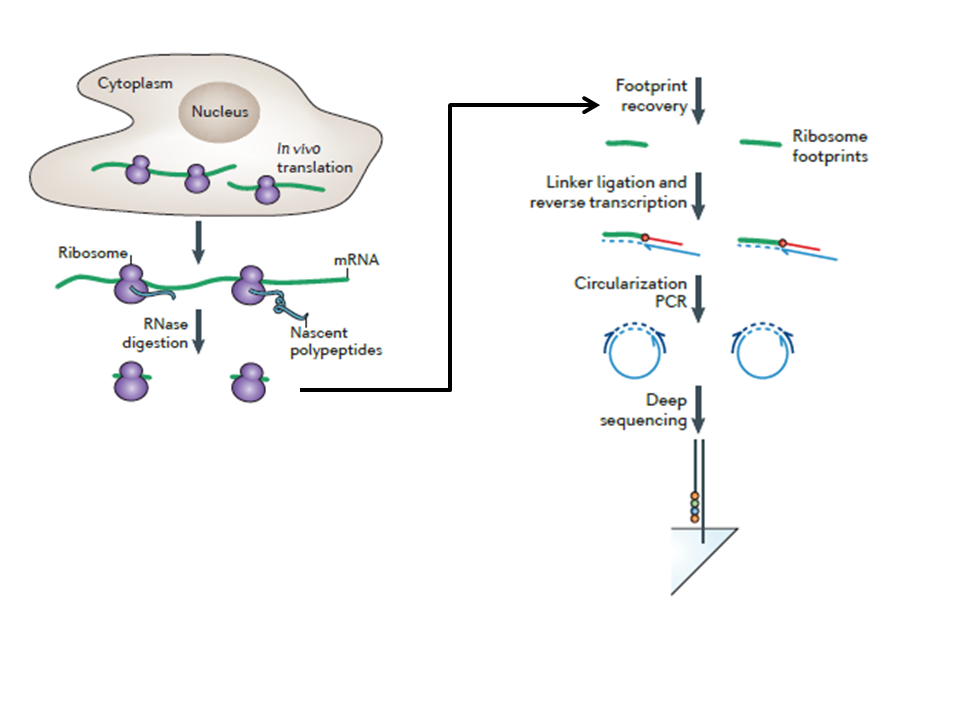
Figure: Diagrammatic representation of Ribosomal footprinting. Source: Ingolia NT et al., Nature Protocol, 2012
Advantages of Ribosomal footprinting:
- It is helpful for following experimental requirements:
- Investigation of Translational control and gene expression
- Spotting translation start sites
- Analyse relative kinetics of protein synthesis and abundance
- It is a fast, efficient and scalable spin column method
- It shares compatibility with different type of samples such as cell lines, mammalian tissues, and yeast






