 As a functional unit of a living entity, cells are very complicated and consist of exceptionally high number of biomolecules with completely different physical and chemical properties. The complexity of the biomolecules profile is even more intricately woven depending upon their subcellular location. With the limited resolution of the separation and localization technologies, it is imperative to employ fractionation methods for protein profiling and expression analysis. Even the lesser abundant proteins can be analysed if screened after prefractionation. Sensitive and accurately fractionated samples help in convolution of methods such as mass spectrometry or in-vitro analysis for the detection of proteins at the level of organelles or transportation compartments.
As a functional unit of a living entity, cells are very complicated and consist of exceptionally high number of biomolecules with completely different physical and chemical properties. The complexity of the biomolecules profile is even more intricately woven depending upon their subcellular location. With the limited resolution of the separation and localization technologies, it is imperative to employ fractionation methods for protein profiling and expression analysis. Even the lesser abundant proteins can be analysed if screened after prefractionation. Sensitive and accurately fractionated samples help in convolution of methods such as mass spectrometry or in-vitro analysis for the detection of proteins at the level of organelles or transportation compartments.
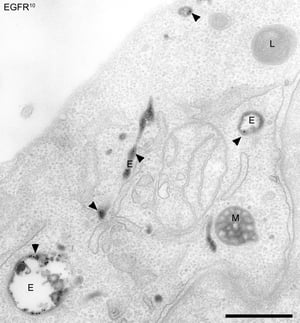
Endosomal organelles are enriched with many proteins that are being transported across the cell for performing vital functions in a compartment-specific manner. Endosomes are membrane-bound vesicular organelles that are crucial for transportation, sorting and synthesis of many macromolecules. Isolation of endosomes provides a lot of information about compartment-specific profiling of the macromolecules.
Compilation of previous studies reflects the spatio-temporal existence of different stages of endosomes such as early endosomes and late endosomes. All these subpopulations vary dramatically in terms of proteomic composition and complexity. Isolation of these endocytic organelles provides a fundamental tool of understanding intracellular signalling events. Over the last few years, various methods were developed to enhance the knowledge about endocytic proteins and their isolation. This blog will discuss briefly about the methods of isolation of endosomes and their further analysis.
Density gradient-based ultracentrifugation
Subcellular fractionation using density gradient based centrifugation provides a great method to isolate endosome organelles and multi-protein complexes.
The overall method has two crucial steps:
- Homogenization: The tissue/cells are homogenized in suitable media to burst open the cell membrane and release the cell organelles and cytoplasmic components into the lysis buffer. The collected cells are briefly centrifuged to remove the cellular debris, unbroken cells, and nuclei. The supernatant obtained at this step consist of cell organelles or membrane bound proteins.
- Ultracentrifugation based fractionation: The post-nuclear supernatant obtained after homogenization can be separated into its components using different gradients followed by ultracentrifugation. Discontinuous gradients and step gradient, both of these methods are applicable for the fractionation. The place where a particular membrane or endocytic organelle can be isolated in the gradients is based on the ratio of the lipid content of the endosomes to their protein contents. Heavier cell organelles such as mitochondria and endoplasmic reticulum have high protein content; therefore tend to settle at the bottom of the gradient. Lipid-rich organelles such as endosomes have low density therefore they are isolated from the middle layers or upper layers of the gradient.
In order to enhance the resolution of the fractionation, the most preferred method for this is using equilibrium separation in a continuous gradient. The post-nuclear supernatant is allowed to equilibrate in a continuous gradient in order to distribute the endosomes of different densities according to their sedimentation quotient. Since the abundance of same density endosome is already low, the only disadvantage of using equilibrium separation is getting a diluted fraction of the endosomes. This drawback can be negated by applying prefractionation approaches such as chromatographic separation, which helps in enriching the sample further.
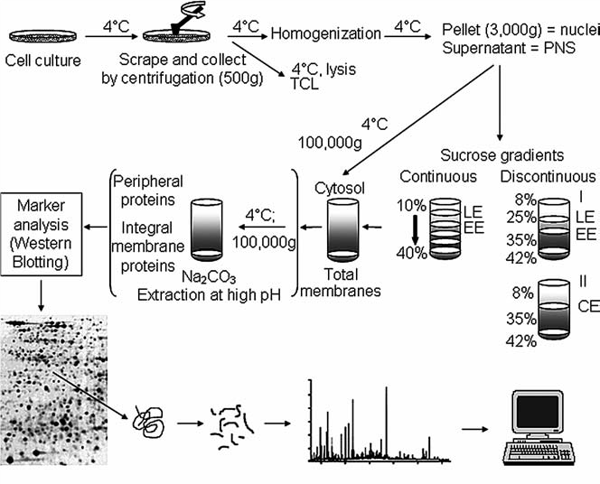 Figure 1: Schematic outline of fractionation and isolation of endosomal organelles using gradient based ultracentrifugation followed by proteome analysis. Source: Araújo et al., Methods in Molecular Biology. 2008.
Figure 1: Schematic outline of fractionation and isolation of endosomal organelles using gradient based ultracentrifugation followed by proteome analysis. Source: Araújo et al., Methods in Molecular Biology. 2008.
Fluorescence-activated organelle sorting (FAOS)
This method utilizes basic principles of flow cytometry to sort and detect the specific endosomes having peculiar light scattering and fluorescent properties. Fluorescent probes, labelled antibodies or fluorescent proteins are used to tag specific organelles. In a way, fractionation is coupled with high speed sorting of the endosomes. The limiting factor in this technique is to decrease the background signal created by the absorption of light by the contents of the sample.
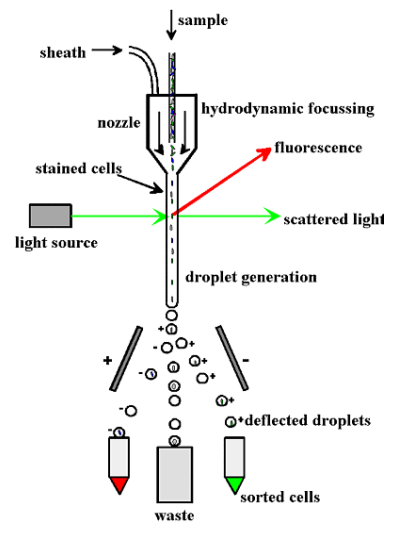
Figure 2: Diagrammatic representation of FAOS. Source: Stasyk and Huber, Proteomics, 2005.
Free Low electrophoresis (FFE) and Flow Field-Flow Fractionation
This method harnesses the property of differential mobilities of different organelles based on their membrane properties when exposed to an electric field followed by collection of the sample at different time points. Principally, the sample is allowed to move in a laminar flow manner and then a suitable electric field is applied perpendicular to the flow. The resultant potential causes the displacement of organelles along the electric field. The final displacement and mobility of the organelles is decided by the surface biomolecules and the net charge on them. This is a great tool to isolate an enriched fraction of the organelles especially the small ones such as endosomes. Each fraction can be used for further analysis based on the organelle specific biomarkers.
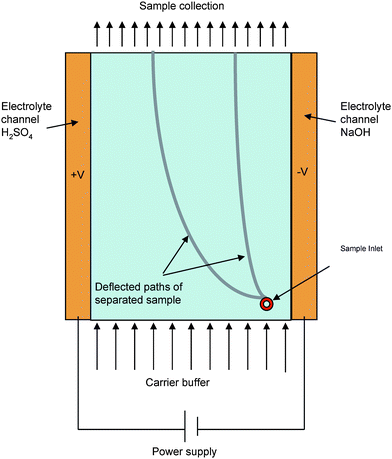
Figure 3: Schematic representation of a free flow electrophoresis. Source: Ho S et al, Journal of Materials Chemistry, 2009.
Immunoisolation of endosomes
This method relies on binding separation of organelles using their antigenic properties rather than using their physical properties, unlike other isolation methods. The basic principle is closely similar to antibodies and antigen binding as any other immunological method. However, the parameters of choosing antigens, antibodies and the support on which it is growing vary substantially.
- Antigens: Immunoisolation works on a one to one ratio which suggests a single epitope is enough for separating a single vesicle. Therefore, the epitope that should be chosen must be exclusively present on the desired endosomal compartment. Since, this method does not consider the variation in physical properties such as density of the antigen; therefore, this method is not suitable for differential immunoisolation. Care should be taken about the accessibility of the selected epitope. It should be easily accessible to the antibody which is immobilized on a solid surface. It is always suggested to select the epitope which has higher abundance to increase the efficiency of binding.
- Antibodies: Antibodies are held on a solid support in this method, therefore two different types of antibodies are used for an efficient binding.
- Linker antibody- it is used to enhance the range of binding and flexibility of the specific antibody. It couples the solid surface beads with the specific antibody to allow endosomal binding. Mostly commonly a generic anti-Fc antibody is used as a linker antibody.
- Specific antibody: These antibodies are raised against the epitope present on the surface of the endosomes that are required to be isolated. It is suggested to use an affinity purified polyclonal antibody. However, the binding of antibody with the endosomes require optimisation and calibration.
- Solid Support/matrix:Binding of the linker antibody on a solid surface is crucial. It is also imperative to choose a solid matrix carefully.
- Flexibility: An ideal matrix should be flexible enough to bind with the antibody without a positional disorientation and conformational hindrance.
- Sedimentation: Matrix should be able to aggregate and sediment easily at a very low speed like 3000 x g at which the endosomes do not co-sediment. It should not be cross linking with the endosomes as well so that both of these can be easily separated after binding.
The most common modification of immunoisolation is the use of Magnetic beads. This approach is beneficial in being rapid, gentle and efficient. It does not require high speed centrifugation or density gradient based fractionation for separation of endosomes. It is also a useful method to isolate differential densities of endosomes with greater efficiency and purity.
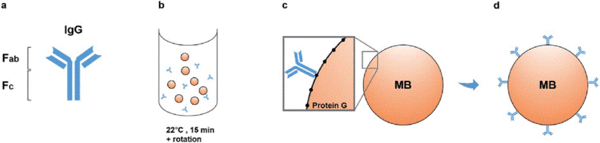
Figure 4: Schematic representation of magnetic beads based immunoisolation. Source: Lordachescu A et al., Royal Society of Chemistry Advances, 2018
Magnetic beads can be used in two ways:
- Direct Binding: The magnetic beads are incubated with the antibody first for a short duration followed by a longer incubation with the crude sample containing endosomes. The targeted endosomes attached with the beads can then be separated by using a magnet and few washing steps.
- Indirect Binding: In this protocol, the antigen and the antibodies are allowed to bind first. Magnetic beads are added with the antigen/ligand mix later on and allowed to bind for some time. The extraction and purification method is similar as that of direct binding technique.
Direct binding technique is recommended when the antigen is easy to access. When the targeted antigen is difficult to access, the indirect binding technique is preferred.
Points to be considered during endosomal isolation:
- Nuclear rupture while homogenisation releases DNA resulting into increased viscosity of the solution. Therefore, enough detergent has to be added to maintain the consistency of the homogenate.
- Cross contamination of the samples should be avoided from the standards.
- Endosomes are very fragile under ionic conditions. Therefore, the homogenisation of the tissue should be very mechanical and mild in order to avoid their rupture.
- While making gradient in the density-gradient fractionation, air bubbles should be avoided as they interfere with the continuity of the gradient.
- The layers after ultracentrifugation should be collected carefully so as to obtain a pure population of the organelles.
- Endosomal population obtained after the method should be checked for purity using endosomal markers.






