Earlier designated as the "garbage bags" used by the biological systems to remove unnecessary biomolecules of the cells, Exosomes, have recently gained popularity as secreted vesicles pivotal for intracellular and intercellular information transfer. Studies have shown the presence of essential RNA and protein cargos in these small vesicles (30-150 nm). The interest in exosome studies have exponentially grown as they can be manoeuvred as minimally diagnostic tools in order to understand biological functions. Proteomics of exosomes are considered the biological fingerprints as they replicate the properties of the parental cell from where they are originated. As a mobile container of vital biomolecules (specialized proteins and RNAs) exosomes are crucial for antigen- presentation, cell-cell communication, waste management, translocation of biomolecules and coagulation.
Moreover, recent evidences have reflected the role of exosomes as a non-invasive diagnostic marker to understand pathological conditions as well as connect anterograde or retrograde communications.
Critical study of isolation, characterization and analysis of exosomes and their RNA and protein content can provide an in-depth probing and information in terms of function. Characterization of exosomes can be done by using metabolic incorporation of labelled/modified nucleic acid such as ethynyl uridine or methionine analogs such as homopropargylglycine. The crucial part of the study is to get a pure fraction of exosomes without any subcellular contamination. Hence, various methods have been developed for obtaining pure exosomal fraction.
The most common approach used for the vesicular isolation is ultracentrifugation- based purifications, but this allows infiltration of non-vesicular components of the cellular system, which becomes challenging for consistent diagnosis from samples such as serum, urine or tissue. The other approach is using electron microscopy to get the contaminants information; however this is not a quantitative approach.
Exosome Isolation Techniques
-
Ultracentrifugation-based isolation methods:
The fundamental principle of this method is to separate the components of the cellular compartments on the basis of the sedimentation quotient (density, size and shape) after application of centrifugal force. The denser particles will settle first. The separation also depends on the physical properties of the particles, density and viscosity of the solvent. Ultracentrifugation uses exceptionally high centrifugal forces up to 1,000,000 × g, which could separate samples based on two ways- analytical and preparative ultracentrifugation, depending upon the protocol used. Exosome isolation by ultracentrifugation is the gold standard method. Preparative ultracentrifugation has two different methods- differential ultracentrifugation and density gradient ultracentrifugation.
The general protocol of differential ultracentrifugation consists of a sequence of centrifugation cycles of different durations and speed ranging from 100,000 to 120,000 × g. The first step of isolation is the cleaning of the sample to get rid of contaminants and the sample is mixed with protease inhibitors to prevent the degradation of exosomal proteins. After repeated centrifugation, the supernatant/pellet is collected and resuspended in a suitable medium such as PBS, which is further subjected to few more runs of increasing centrifugal force, till the final exosomal fraction is obtained. The Final fraction is resuspended and can be used for further analysis.
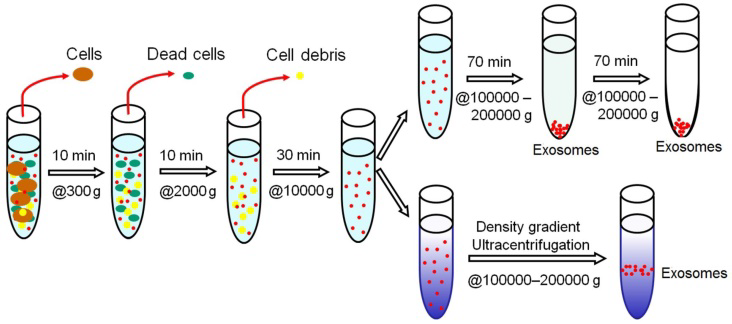
Figure: Schematic representation of Ultracentrifugation-based Exosome Isolation. Source (Pin Li et al., Theronostics, 2017)
Density gradient ultracentrifugation separates exosomes based on the size and density of the vesicles in a pre-constructed gradient medium in a tube (progressively decreasing density Top-to-Bottom). Firstly, the sample is layered on the top of the gradient, followed by multiple cycles of ultracentrifugation separating the exosomal fraction at a specific gradient layer. Continuous gradient is useful for analytical purposes while discontinuous gradient is preferred for preparative requirements.
-
Size-based isolation techniques
This method utilizes the conventional membrane filtration where the separation of the components is principally based on their size or molecular weight. Hence, the exosomes can be extracted using the membranes within their size exclusion limits. Comparatively, it is faster than ultracentrifugation and easy. Specialised nanomembranes can be used for exosome isolation and diagnostic purposes. Commercially available exosome isolation kits are syringe filter-based rapid fractionators which can be used for the separation of exosomes from cell-free samples like urine, serum, cerebrospinal fluid, and cell culture medium. In the syringe filter cartridges, two membranes are tandemly located in close proximity with each others in a way so that, the lower membrane captures small vesicles such as exosomes while the larger components stays at the top membrane.
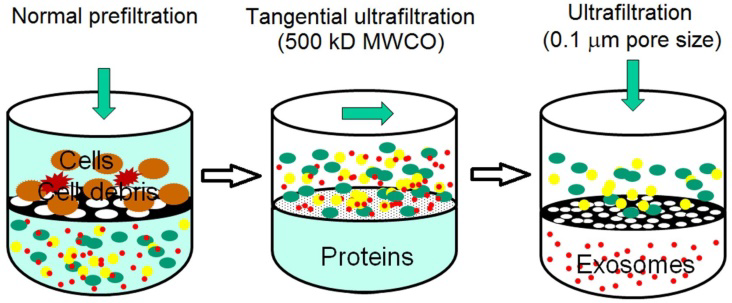
Figure: Diagrammatic illustration of ultrafiltration. (Source: Heinemann ML et al., J. Chromatogr. A, 2014)
-
Immunoaffinity capture-based methods
Immunoaffinitive interactions of the proteins and receptors in the membrane of exosomes with their antibodies provide wide opportunity for the development of specific techniques of exosomal isolation and diagnosis of specific receptors. Various modifications have been developed so far for immuno-isolation of exosomal biomarkers and their sources. Microplate-based enzyme-linked immunosorbent assay (ELISA) is one of the methods for isolation and quantification of exosomal secretion in plasma, serum and urine samples. These results can be interpreted as absorbance readouts compared with the known surface biomarker`s expression, which enables the specificity and accuracy of this method.
Mechanistically, the first step is pre-cleaning of the samples using pulses of low-speed centrifugation, avoiding large particle contamination. Submicron-size magnetic particles may enhance the efficiency of immunoisolation. Even with a little amount of sample (1 ml of supernatant), the yield is higher. With plasma samples, the yield is 10-15 folds higher than ultracentrifugation.
-
Microfluidics-based isolation methods
Fabrication of microfluidics-based isolation devices are greater advancement in the field of exosome isolation utilising the biophysical and biochemical properties of the exosomes at microscale level. Apart from the conventional approaches of size, density and immunoaffinity based sorting, many other modifications can be used such as acoustic, electrophoretic and electromagnetic manipulations, enhancing the efficiency, isolation time and sample requirement.
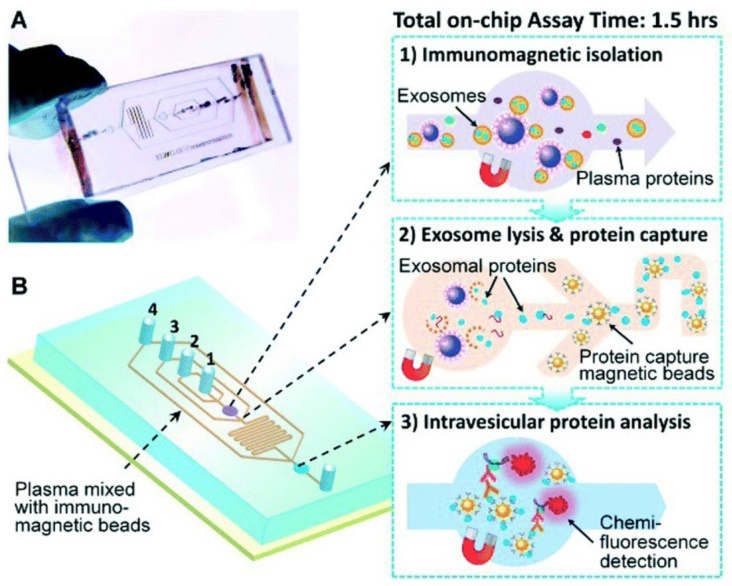
Figure: Schematic representation of microfluidic chip based isolation of exosomal proteins (Source: He M et al., Lab Chip 2014)
Which method to choose for exosomal isolation?
|
Methods |
Pros |
Cons |
|
Ultracentrifugation based methods |
Larger sample capacity with higher yield Reagents are cost effective |
Tedious, long run protocol with labor intensive requirements. High speed may damage exosomal fraction |
|
Size-based methods |
Ultrafiltration is faster, portable and safer No requirement of special equipment Highly reproducible |
Compromised purity Sheer induced degradation Loss of exosomal fraction in the membrane |
|
Immunoaffinity capture-based methods |
Higher purity Isolation of specific exosomes Sub-typing is feasible |
Reagents are costly Requires specific exosome tags Lower yield ·Works well only for cell-free samples Antigenic epitopes may not be exposed |
|
Microfluidics based methods
|
Low cost devices Higher portability Automated and easy to integrate |
Still in the process of validation and standardization Low sample capacity |






