Nature has ability to generate an unending array of molecules from very small number of building blocks, even simple microbes such as E. coli have synthetic chemical prowess that can bring the most sophisticated organic chemistry labs to their shame. Inspired by the nature’s ability to knit complex molecules from a small number of building blocks, using simple modular reactions, Berry Sharpless, and his coworkers, defined and invented the area of ‘click chemistry’. Together, Berry Sharpless and M. G Finn redefined the use of simple modular reactions that had already existed to generate new ‘functions’ or properties and termed it as ‘click chemistry’.
What is click chemistry?
For a chemical reaction to be included as a click reaction it must have following essential features:
- Reaction should be modular with no side products.
The key fact is ‘Click’ reactions do have a high thermodynamic driving force. There is just one predominant outcome of reaction, like fitting blocks together in a LEGO with a “click” you get two simple molecules combine. - Should be easily performed in benign solvents like water
Click chemistry is green in its approach - No protection and deprotection is required.
Simple, hassle free organic synthesis is the aim. - The product should be easily isolated; no chromatography is required.
Many existing chemical reactions fulfill the criterion of being a click reaction such as Cycloadditions; 1,3-dipolar cycloadditions, Nucleophilic ring-opening reactions; refer to the opening of strained heterocyclic electrophile. Moreover, non aldol carbonyl chemistry (aldol carbonyl reactions generally have low thermodynamic driving forces, hence they lead to formation of by-products and reaction yield is low and therefore not a click chemistry reaction by definition) and addition to carbon-carbon multiple bonds, dihydroxylations, epoxidation, Michael addition etc are also click reactions.
The most popular and widely used click reaction however, is Cu(I)- catalysed Huisgen, 1,3-dipolar cycloaddition reaction of terminal alkynes and azides to generate 1,2,3-triazoles (See Figure 1A). Copper catalysed Huisgen reaction can happen in aqueous medium, can tolerate a wide range of pH from 5 to 12, it does not require any thermal activation (can occur between 0 to 160°C). The most interesting part about this click reaction is its orthogonal character, which has made it so popular amongst biochemists, chemical biologists and protein chemists. The term orthogonal nature or bio-orthogonal character implies that the participating reactants, a terminal alkyne and an azide moiety, can combine with each other, driven by a high thermodynamic force but they don’t react with other molecules in the cell, which according to Barry Sharpless is “is to create highly reactive ‘sticky spots’ with the right target groups while having them remain invisible to most other types of molecule”. This character is called bio-orthogonal and enables chemical biologists and protein scientists to apply this reaction to a range of applications for chemical proteomics.
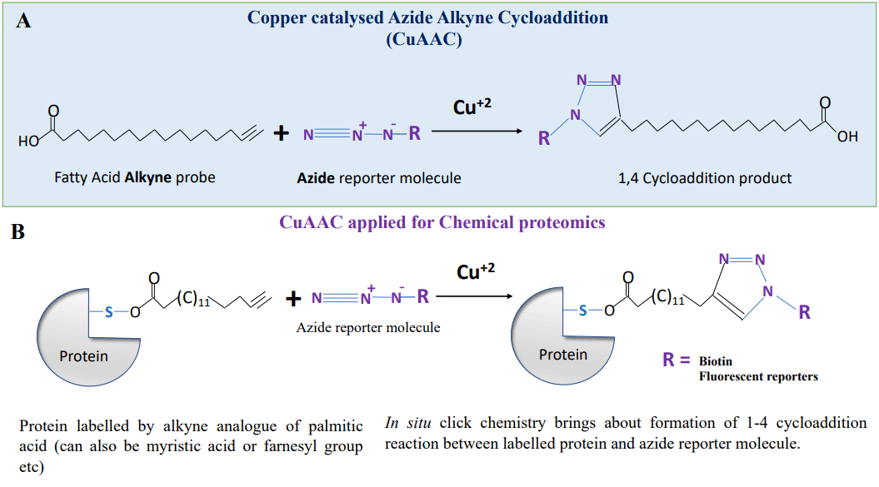
Figure 1. Copper catalysed azide alkyne chemistry (A) the Huisgen reaction (CuAAC reaction), an azide and alkyne molecule combine together to make 1-4 regioselective cycloaddition product which is 1,2,3 triazole (B). CuAAC reaction applied to chemical proteomics for studying post translational modification of proteins by acyl moieties.
Applications of Click chemistry to proteomics
Biologists have found many creative applications of Huisgen or CuAAC reaction. To use CuAAC chemistry in cells, azide and alkyne derivatives of biological molecules are required. These molecules are known as clickable analogues of corresponding molecules, because they can undergo Huisgen cycloaddition reaction (Figure 2A; 2,3). Some of the prevalent example of application of click chemistry in biology are following.
BONCAT : Capturing Nascent proteome by CuAAC chemistry
BONCAT refers to Bio-orthogonal non-canonical amino acid tagging. It has been applied to study dynamic changes in protein translation in cells. BONCAT requires labeling cells with a modified amino acid, Azidohomoalanine (AHA), this is an azide derivative of methionine, which is non-toxic to the cells. The translation machinery cannot distinguish between AHA and methionine and exogenously supplied AHA is incorporated by cells in newly synthesized proteins in place of methionine (Figure 2B). The azide function on azidohomoalanine provides a ‘sticky spot’, which does not react with other molecules inside the cell, but can only be conjugated with a clickable reporter molecules containing alkyne function in presence of copper sulfate (Figure 2C). Such alkyne reporter molecules can be alkyne derivatives of biotin or alkyne derivatives of fluorescent molecules. The term non canonical refers to non-standard clickable amino acids. The incorporation of clickable amino acid, AHA in cellular proteins enable protein scientists to distinguish the newly synthesized proteins from pool of proteins in the cell. Very often, cells respond to a given stimulus by translation of new proteins, such as response to cytokines, response to steroid hormones, response to infection, response to ligand receptor interactions etc, these proteins are a part of a global program required to adapt cell to a given stimulus. BONCAT provides a powerful way to profile newly synthesized proteins in cell.
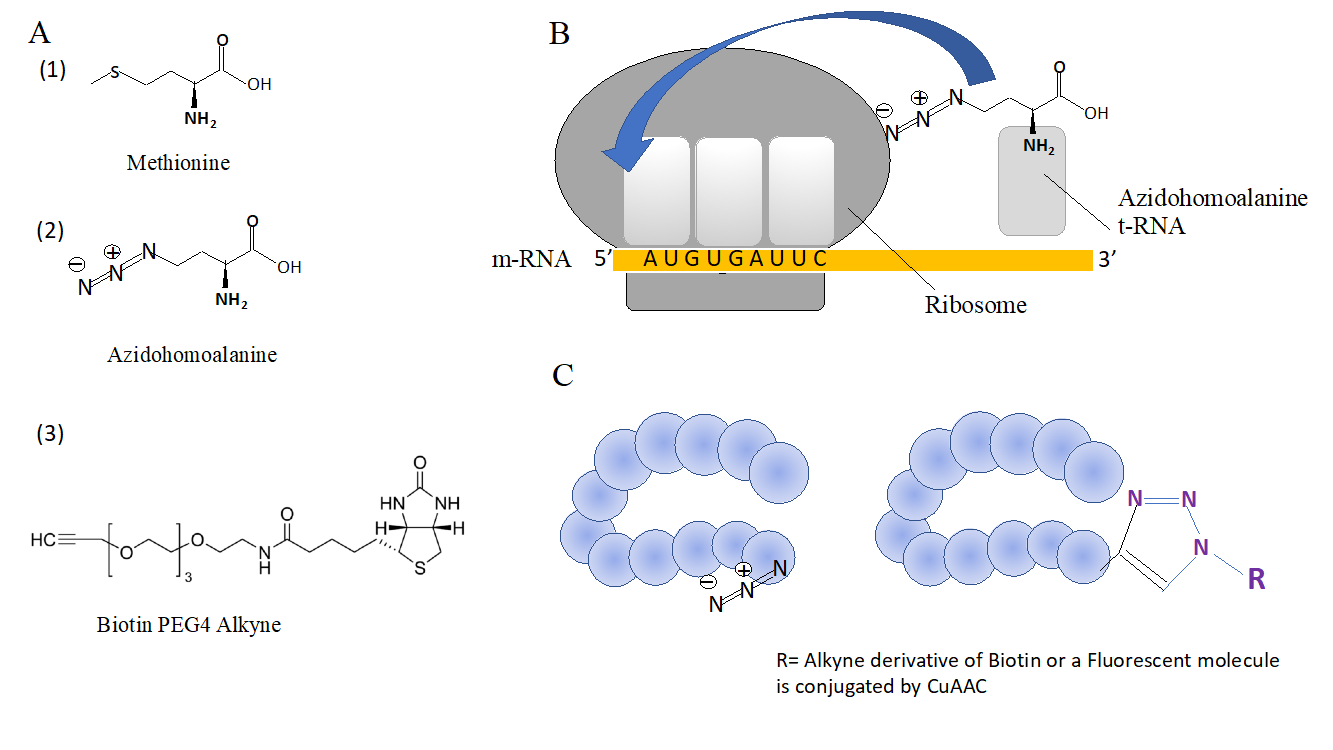
Figure 2. Application of CuAAC for chemical proteomics.(2A), Molecular structures of (1)methionine and (2) Azidohomoalanine (clickable analogue of methionine) and Alkyne derivative of Biotin (3); (2B) The translational machinery of cell incorporating AHA in growing polypeptide; (2C) Proteins labelled with AHA undergoes CuAAC to form 1,4 cycloaddition product combining a reporter alkyne molecule to the azide of AHA.
Metabolic labeling and CuAAC to study PTMs
A large number of clickable analogues have been designed and are now commercially available to study the post translational modifications, such as farnesylation, myristoylation, glycosylation and palmitoylation. These clickable molecules can serve as analogues or surrogates of cellular molecules that modify the proteins for these PTMs. The clickable analogues are supplied exogenously in the culture medium to the cells and they compete with their natural counterpart and gets incorporated into the proteins. Similar to BONCAT, this provides a provision to distinguish the newly post translationally modified proteins from the global pool of post translationally modified proteins. Figure 1B illustrates a protein post translationally modified with clickable analogue of palmitic acid, which can be conjugated with biotin alkyne or with a fluorescent reporter molecule to render the palmitoylated proteins fluorescent. The Bio-orthogonal nature of clickable probes provides ability to monitor the post translational response of a given type to a given stimulus in cells. Many PTMs are very dynamic and application of metabolic labeling of cells with clickable analogues allows for monitoring the dynamic response of such PTMs.
CuAAC chemistry has many other applications in polymer industry, medicinal chemistry and activity based profiling of enzymes. Click chemistry is simple, elegant and yet a powerful approach. The CuAAC reaction is extremely simple and anyone can do it easily in situ. Click chemistry has revolutionized the way chemical proteomics is done in lab these days.
References
- Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. (2003) Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003 Mar 19;125(11):3192-3
- Hein CD, Liu XM, Wang D. Click chemistry, a powerful tool for pharmaceutical sciences. Pharm Res. 2008 Oct;25(10):2216-30.
- Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem Biol. 2009 Jul 17;4(7):581-7.
- Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR, Heal WP, Broncel M, Serwa RA, Brady D, Mann DJ, Leatherbarrow RJ, Tewari R, Wilkinson AJ, Holder AA, Tate EW. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat Chem. 2014 Feb;6(2):112-21.
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bio-orthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci U S A.2006 Jun 20;103(25):9482-7.






