.png?width=300&height=185&name=new_picture_(10).png) Since 1975, immobilized metal affinity chromatography (IMAC) has been popularly used in purifying proteins, especially those that are fused to a polyhistidine tag, typically a 6X His tag. This process gained immense popularity since it allows for the efficient purification of proteins, even those from crude lysates. In addition, its robust nature makes it ideal for methods that require protein-specific conditions. Its functional simplicity, affordability and compatibility with a wide range of reagents also add to its popularity.
Since 1975, immobilized metal affinity chromatography (IMAC) has been popularly used in purifying proteins, especially those that are fused to a polyhistidine tag, typically a 6X His tag. This process gained immense popularity since it allows for the efficient purification of proteins, even those from crude lysates. In addition, its robust nature makes it ideal for methods that require protein-specific conditions. Its functional simplicity, affordability and compatibility with a wide range of reagents also add to its popularity.
Understanding IMAC: How Does the Process Work?
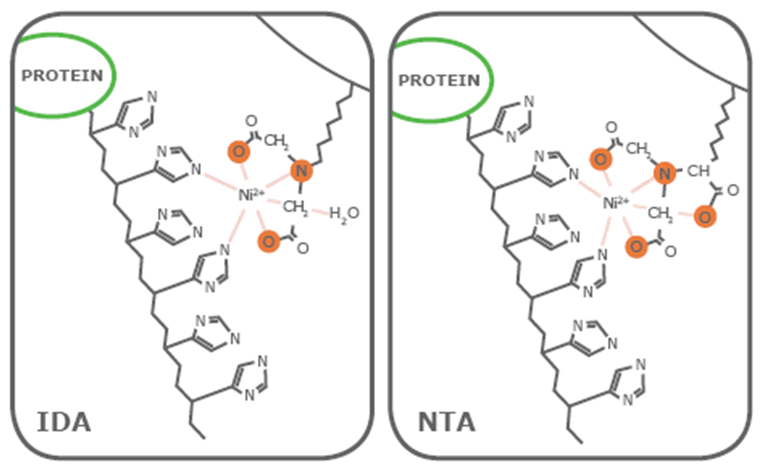
In a nutshell, the polyhistidine tag and bound protein are sequestered from the sample solution by using affinity resins. Transition metal ions fixed to a matrix through a chelating ligand such as nitrilotriacetic acid (NTA) and iminodiacetic acid (IDA) interact with imidazole rings of the polyhistidine tag and isolate them, along with the fused protein, from the sample solution. The fused protein can then be dissociated from the metal ion through any one of these methods – elution with an imidazole gradient, metal chelation or changes in pH.
IDA vs. NTA: Which One Should You Use?
Let us start by saying that the yield and purity of your eluted protein heavily depends on the particular ligand that you will use in the process. Generally speaking, the number of valencies with which the ligands coordinate the metal ions significantly affects the quality of the purified protein.
Iminodiacetic acid (IDA) and nitrilotriacetic acid (NTA) are two of the most commonly used IMAC affinity resins for protein purification. While both ligands have two valencies available to interact with the histidine residues, IDA has a total of three valencies available while NTA has four. Now, since we have earlier said that the number of valencies affect the overall quality of the purified protein, the additional carboxymethyl group present in NTA makes it a stronger coordinator of metal ions.
Being a trivalent ligand, IDA-based resins leach metal ions at a higher rate, particularly in the equilibrium and wash steps, as compared to NTA-based resins. It was also observed that the purity of His-tagged proteins eluted using an IDA-based resin is significantly lower as compared to those from an NTA-based resin.
Despite these limitations, however, IDA-based resins are still widely popular due to the following reasons:
-
They are less expensive.
-
They require a lower imidazole concentration to elute proteins.
-
They provide higher metal loading capacity to generate greater yields.
So, how do you really choose which resin to use? While most researchers cannot give a definite answer to this question, you need to consider the fact that the binding capacity of an IMAC resin is heavily dependent on the nature of the protein being purified and the metal ion used in the process.
For example, IDA may exhibit a higher metal ion capacity and avails more binding sites for a target protein as compared to NTA but you also need to consider the fact that the binding capacity differs from one protein to another. In addition, the metal ion used in the process also helps to determine the results of the experiment. Cobalt-based resins have high binding specificity but exhibits low affinity while nickel-based resins have higher affinity but lower binding specificity.
The presence of compounds such as DTT (a reducing agent which stabilizes proteins with free sulfhydryl groups) and EDTA (a hexadentate chelating agent found in most buffers) in your protein samples can also affect the binding capacity of IDA and NTA-based resins.
Taking all of these things into consideration, you may need to explore different metal ion and ligand combinations to get the best results from the purification of your His-tagged proteins. You also need to make sure that your study samples are free from compounds that may affect the results of your experiment.






