In recent years, powerful tools to disrupt gene and gene products have were devised; some of these tools allow conditional disruption of either the DNA or mRNA corresponding to a given gene. Such tools allow interrogation of gene function with unprecedented precision, Control and flexibility. Conditional silencing of gene function is a powerful tool for geneticists and cell biologists. It enables interrogation of the role of ‘essential’ or ‘quasi-essential’ genes that cannot be deleted altogether from a genome.
However, all the methods of conditional-gene disruption target precursor steps, such as deletion of DNA or mRNA transcripts, have one critical limitation: the delay in the manifestation of the phenotype after the disruption. This is because all these methods are pre-translational, i.e. happening before the protein translation and not directly acting on proteins. And even after disrupting some degree of phenotype exists transiently due to existing population of protein molecules for the disrupted gene/mRNA. Wandless and co-workers found that a post-translational approach to regulate gene function is one of the most effective and fastest ways to study the effect of perturbation. This is because the perturbation acts directly on the proteins.
But how to achieve conditional disruption of protein molecules corresponding to a given gene? Within the living cells, proteins are dynamic entities and are subject to cycles of synthesis and degradation; each protein has a specific half-life. Unstable or misfolded proteins are degraded by proteasome machinery of the cell. The scientists have capitalized on this fact; they designed a contrivance for conditional destabilization of a protein for its degradation when not required. In this way, the protein molecules encoded by a given gene can be stabilized or degraded in a tunable fashion.
By appending an unstable version of DHFR protein (from E. coli) to the target protein's terminus, the target protein also become unstable and susceptible to proteasomal degradation. However, the unfolded structure of DHFR can be stabilized with Trimethoprim (TMP), a competitive inhibitor of DHFR. The stabilization of DHFR secures stability of the target protein to which it is appended. In the absence of TMP, DHFR protein is not stabilized, and destabilized DHFR and target protein is rapidly degraded by proteasomal machinery of the cell (see Figure 1)
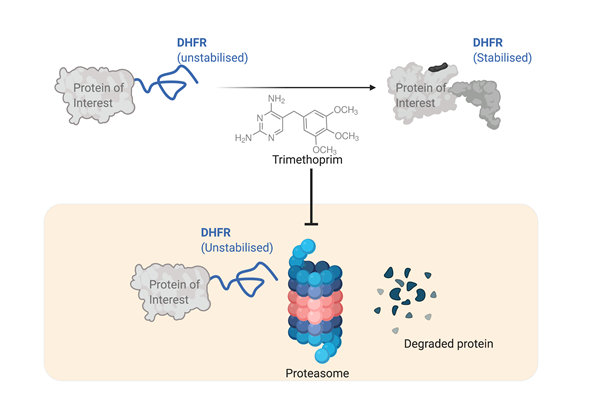
Conditional destabilization of mammalian proteins using an inherently unstable DHFR. The unstable DHFR (unfolded, see Figure) is rapidly degraded along with the associated protein. The stability of DHFR is tuned by presence of TMP (Trimethoprim). Thus, TMP stabilizes DHFR and also the associated protein.
DHFR is an E coli dihydrofolate reductase that catalyzes the reduction of dihydrofolate to tetrahydrofolate. The E. coli DHFR can be targeted by TMP more potently than its mammalian counterpart allowing a large therapeutic window for the use of TMP in mammalian systems. The protein of interest can be tagged with DHFR using DNA transfection with DHFR cassette.
The DHFR-TMP system is used successfully to destabilize many diverse proteins in mammalian cells. The DHFR system can act in an orthogonal manner to already existing FKBP/Shield system (will be discussed in some other blog article), allowing both approaches simultaneously. This can enable simultaneous conditional destabilization of two genes at a time. However, the FKBP/Shield system suffers from a significant drawback. It cannot be used to target transmembrane proteins. This limitation is overcome in the DHFR system where transmembrane proteins can also be knocked down efficiently.
From a pharmacological standpoint, TMP can cross difficult physiological barriers such as the blood-brain barrier and placental barrier, allowing this system to tune protein function in otherwise non-amenable targets such as the mammalian brain and embryonic development.
The DHFR system allows conditional chemical knockdown of genes at protein level (post-translational), allowing critical genes' study indispensable for growth and development refractory to permanent deletion. A conditional approach using either DHFR-TMP or FKBP-Shield allows rapid and tunable Control of gene function compared to other systems.
References:
- Iwamoto M, Björklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010 Sep 24;17(9):981-8. DOI: 10.1016/j.chembiol.2010.07.009. PMID: 20851347; PMCID: PMC2943492.






