Transcript analysis and DNA information is a crucial step in the field of Molecular biology and genetics. Since many years’ DNA hybridisation is utilised as a primal technique for diagnosis of genetic manipulations and their correlation with the diagnostic manifestations of the genetic diseases. The identification of DNA or RNA has become pivotal in many clinical studies and drug development approaches.
The most recent development in this field is a new class of oligonucleotide probes known as Molecular Beacons (MB). Since their development, MBs show wide use in different areas of detection of single nucleotide polymorphism (SNPs), biochip development, wide-scale genetic screening, mRNA-tracking in living systems and development of biosensors. The agility and free modification range allow these sensors to be used in the interaction with DNA, RNA and proteins.
Structure of a standard Molecular Beacon:
MBs are single-stranded small oligonucleotides having specialised structures:
- A target-recognition region (15-30 bases)
- Two short complementary stem sequences flanking the recognition site
Combinatorically, this entire arrangement provides a very conventional stem-loop structure in native state. Each stem sequence end has a fluorophore and a quencher which are in close proximity with each other resulting into effective quenching of the fluorescence through energy transfer when the MB is not attached with any target. The loop portion is specially designed to complement the target site emphasizing on the specificity of the molecular beacon. The spatial orientation of the quencher and fluorophore provides thermodynamically stable confirmation and also enable an efficient and flexible intrinsic switching of fluorescent and non-fluorescent states giving an exceptional advantage of selectivity, sensitivity and real-time detection of the hybridization.
These properties of the MBs are deduced by two working parameters:
- Incomplete Quenching: The baseline or residual fluorescence intensity in the stem-closed confirmation of the MBs.
- No Quenching: Complete stem-open form having resultant fluorescence intensity.
In principle, the complete stem-closed state should not have any fluorescence at all as the quencher ideally quenches the fluorophore completely. However, there is always a residual fluorescence which greatly affects the efficiency and sensitivity of the detection.
The residual fluorescence can be limited by carefully tailoring the following parameters:
- Identification of most compatible fluorophore/quencher pair
- Chemical Synthesis of the MB
- Attaching method of the fluorophore/quencher groups to the oligonucleotide
Mode of Action of Molecular Beacon:
The modus operandi of a molecular beacon lies under the conformational changes that occur after the hybridization of the MB with a target. The stem of the MB is specially designed for a specific target and it's partially or fully complementary to the target. As the probe comes in touch with a target molecule, it makes a hybridised state with it which causes the quencher and the fluorophore to separate resulting into the restoration of the fluorescence. Therefore, there is emission of an intense fluorescent signal when the molecular beacon is in hybridisation stage.
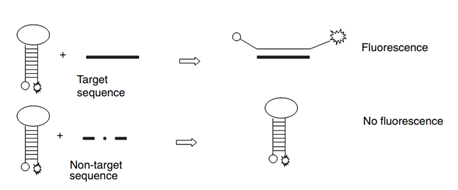
Figure 1: Binding of a molecular beacon with the target or analyte. Source: Goel G et al, Journal of Applied Microbiology, 2005.
Molecular beacon has displayed very high sensitivity even in the identification of changes at the level of single base-pair mismatch. The intensity of signal can be detected easily without separating the probe from the target and is not affected by unbound probe or the target. The binding of the MB with the target is reversible resulting into a dynamic readout.
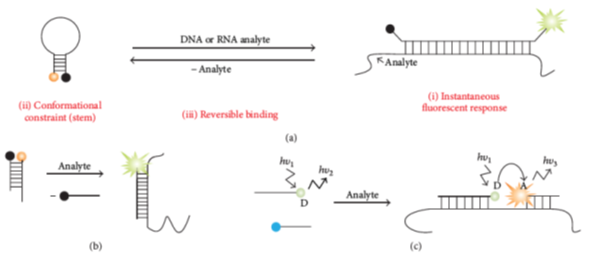
Figure 2: Molecular beacon hybridization. a) Classical probes with binding based on conformational changes happen after binding of a probe with its target/analyte (b) Strand displacement probes (c) Adjacent probes.
Source: Kolpashchikov D M et al, Scientifica, 2012.
Types and properties of Molecular Beacon
|
Class of Molecular Beacons |
Features |
|
Peptide Nucleic Acid-MBs |
Ø Less soluble in an aqueous solution Ø Higher affinity to the analyte Ø High efficiency in terms of binding Ø Resistant to nuclease digestions |
|
Locked Nucleic Acid-MBs |
Ø Very high structural stability Ø Highest affinity to the target than other MBs Ø Higher signal: noise ratio in the living cells Ø Resistant to Nuclease digestion, RNase H and SSBs |
|
Quantum Dot -labelled MBs |
Ø Produce brighter signal Ø Helpful in multiplexing as multiple QDs can be excited at single wavelength |
|
2-OMe-modified MBs |
Ø Higher affinity with the target Ø Sensitive to Nuclease digestions Ø Resistant to SSBs (single stand binding proteins) |
|
Super-quencher MBs |
Ø Can be purified easily Ø higher signal enhancement |
|
gold-Nanoparticle MBs |
Ø High selectivity Ø High quenching efficiency |
|
Conjugated Polymer-linked MBs |
Ø Very bright signal |
Applications of Molecular Beacons:
- Real-time intracellular monitoring:
- mRNA detection in the living cells
- Gene expression studies in the cells
- DNA/RNA biosensors
- Bacterial detection
- Study of DNA and protein interactions
- Single nucleotide polymorphism detection
- Multiplexing
- End point genotyping
- In-vitro detection





