Since the development of cell culture techniques, many modifications are being added regularly to make the conditions controlled on a micro-environment level. With the transitioning research towards drug discovery, it has become imperative to study the mechanisms of cellular development and the responses at a “micro level” regulated both at spatial and temporal switches.
The problems that were usually difficult to address by conventional 3D cultures were the inability to mimic complexity, vasculature of the tissue, spatiotemporally tightly controlled medium exchange as in a in-vivo system.
The most recent advancement in the field of in-vitro studies is using Microfluidics in 3D cultures which gives a window of manipulation of the environment within an artificially microfabricated system. Microfluidic 3D cultures provide many crucial advantages over conventional 3D cultures:
- Spatially controlled co-cultures, allow precise control of molecules with respect to space
- Dynamic control over gradients of molecules (fluidic environment in the channels)
- Integrated approach towards microperfusion control at the level of space and time
A simplified microfluidic chamber harnesses the use of hydrogel-based (or coated) micro-compartments for growing cell spheroids or organoids connected with each other through channels. The viscosity and the flow rate of solutions or media are adjusted according to the need, making it an extremely regulated niche.
|
Properties |
Conventional 3D culture |
Microfluidic 3D culture |
|
External milieu |
Rigid, any change leads to a global difference |
Very flexible, manipulation can be done in an isolated manner as well |
|
Growing conditions |
Standardized pH, CO2, and O2 levels |
Non-standard conditions. They need optimization depending on the experiment |
|
Manipulation level |
Rigid |
Single-cell level manipulation is possible |
|
Perfusion |
Stagnant media |
The perfusion is dynamic and can be controlled spatio-temporally |
|
Reagent consumption |
Large |
Less as compared to macro-cultures |
|
Experimental Analysis |
Endpoint analysis |
Downstream analysis is possible at each step |
|
Sample size |
Requires many cells to seed to establish a proper culture |
Low number of cells is sufficient |
|
Architecture |
Fixed |
Flexible |
|
Cellular resolution |
Low |
Single-cell level resolution can be achieved |
Conventional 3D have some lack when it comes to studying processes like cell migration, scratch or wound healing assays, and growth assays in cells. The problem comes here is the paucity of resolution at a single cell level. Conventional methods require far more work, time and cell seeding, although these experiments can be easily set up and allow live-cell imaging. The loophole here is the difficulty to understand the chemical gradient of molecules and their movement along with it. This problem can easily be tackled by using compartmentalized microfluidic chambers which allow experimental flexibility, time-lapse imaging, and real-time on-chip analysis.
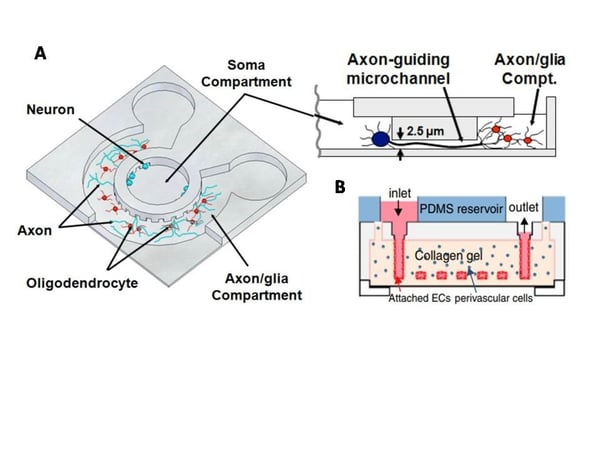
Figure: A: Schematic illustration of the microfluidic compartmentalized CNS neuron co-culture platform (Source: Park et al., Biomed Microdevices. 2009). B: Vasculature in vitro model (Source: Zheng et al., Proc Natl Acad Sci U S A. 2012)
Microfluidic devices provide an extremely important advantage here, which is the precise control over all the conditions on a chip, which on a contrary if done in conventional 3D cultures, require spacious automation and architectures. In a nutshell, it is like a miniaturized version of analytical platforms or a lab-on-a-chip.
For using microfluidic cell culture studies and cellular response certain key parameters are crucial such as:
- Culture material:
PDMS or polydimethylsiloxane is a derivative of the cross-linked polymer of silicone, most commonly used for making microfluidic devices. This polymer is innately flexible, low cost, optically permeable, and porous enough to allow gaseous molecules to pass through them. PDMS allow fabrication of complicated biocompatible fluidic circuits with a very simple prototype. This material can be moulded into a microfluidic device by a technique called Soft lithography. Despite a glamorous use of PDMS in making the chambers, not all cells grow so well on this elastomer. Different modifications of PDMS and other synthetic surfaces are being introduced these days to allow all cell types grow in the microfluidic device.
Studies have revealed that PDMS is an excellent candidate for making an optimal surface for microfluidic devices given the situation where it is cured completely so that the uncross-linked oligomers does not leach out into the culture media.
The surface of PDMS despite all the other favourable properties is hydrophobic, therefore requires treatment so that the cells can adhere to it. There are different ways to make them conducive to cell growth. The common ways are UV treatment, Oxygen plasma treatment, and coating with extracellular matrix proteins such as collagen or laminin, hydrogels, and charged molecules such as poly-d-lysine.
- Diffusion of hydrophobic molecules:
As described in the properties of PDMS, the porosity allows movement of small hydrophobic molecules from the media into the polymer. The diffusion rate depends on the hydrophobicity of the molecules, therefore experiments, where the role of these fat-soluble molecules such as vitamins or hormones is an important parameter, should be optimized well by knowing the diffusion rates. A coating on a PDMS surface can help in reducing absorption by a greater extent.
- External factors:
The external factors such as CO2, O2 level, pH, osmolarity and nutrient circulation are crucial for the optimal growth of cells in a culture. PDMS is extremely permeable to gases which is an advantage but becomes a disadvantage as water vapour gets evaporated continuously apart from permeation of CO2 and O2 to make the media ambient. This evaporation might change the osmolarity by many folds as the surface area is very small. Therefore the cultures need to be buffered continuously with buffers of pH 7.4 to maintain optimal pH and osmolarity. For this purpose, syringe pumps are often used to precisely control the influx and outflux of the buffered media.
Overall, microfluidics devices are much closer to the in-vivo systems with a high level of dynamics control than conventional 3D cultures.






