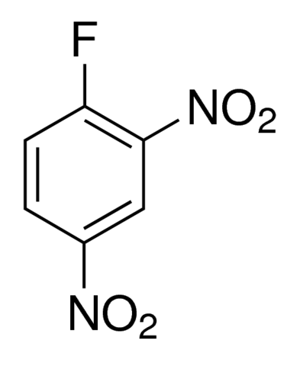
 1-Fluoro-2,4-dinitrobenzene (DNFB), also known as Sanger’s reagent, was first used by Sanger to detect free amino acids of Insulin. DNFB undergoes nucleophilic aromatic substitution with the N-terminal amino group of a peptide or protein. After hydrolysis of the peptide or protein, the individual amino acids separate and only the labeled N-terminal amino acid can be detected by a colorimetric detection at specific wavelength. DNFB is hence used in protein sequencing to determine N-terminal amino acid.
1-Fluoro-2,4-dinitrobenzene (DNFB), also known as Sanger’s reagent, was first used by Sanger to detect free amino acids of Insulin. DNFB undergoes nucleophilic aromatic substitution with the N-terminal amino group of a peptide or protein. After hydrolysis of the peptide or protein, the individual amino acids separate and only the labeled N-terminal amino acid can be detected by a colorimetric detection at specific wavelength. DNFB is hence used in protein sequencing to determine N-terminal amino acid.
DNFB also alters enzyme activity of some proteins. DNFB strongly inhibits the reductase activity of mitochondrial cytochrome b-c1 complex. Addition of DNFB modifies amino groups and basic nucleophilic groups in amino acid residues, resulting in a decrease of both electron transfer and proton translocation in the b-c1 complex. Catalytic properties of Fructose 1,6-diphosphatase are changed by adding DNFB at neutral pH compared to that at high pH. DNFB can also be used to distinguish reduced and oxidised forms of glutathione and cysteine in biological systems especially blood, cell lysates.
Detection of N-terminal Amino acid of Proteins or Peptides:
The N terminal amino acid of the protein is labeled by Sanger’s reagent. DNP derivatives are stable to acid hydrolysis. The labeled protein is then hydrolyzed and loaded on chromatography column and the elution profile is matched with the standard profile obtained from FNB derivative of all the amino acids. The reagents produce colored derivatives which can be easily detected by absorbance. Detection of N-terminal amino acid of Insulin is mentioned below, this method can be applied to any protein (best works with less than 50-70 amino acids.) For larger proteins, before DNFB treatment, the protein should be hydrolysed to small fragments by trypsin or pepsin or by chemicals like cyanogen bromide.
Reagents required
- DNFB
- Insulin
- Sodium bicarbonate
- Ethanol
- Ether
- 20% HCl
Procedure
Preparation of DNP-Insulin
- Solution A- Dissolve 0.5g Insulin and 0.5g NaHCO3 in 5ml water.
- Solution B- Add 0.5ml DNFB to 10ml Ethanol.
- Both Solution A & B are mixed gently for 2 hours at room temperature.
- DNP-Insulin is precipitated as an insoluble yellow powder. This precipitate is washed with water, ethanol followed by ether and air-dried.
Identification of N-terminal amino acids of Insulin
- Mix 100 mg DNP-Insulin with 10ml of 20% HCl and boiled under reflux for 8hr to hydrolyze the protein.
- Let the solution to cool and extract thrice with ether. Ether extract contains ether soluble DNP derivatives (DNP-leucine, DNP-isoleucine,DNP-valine, DNP-phenylalanine, DNP-methionine, DNP-proline, DNP-alanine, DNP-tryptophan, DNP-glycine, DNP-threonine, DNP-serine, DNP-glutamic acid, DNP-aspartic acid, DNP-hydroxyl proline, diDNP-cystine, diDNP-lysine, diDNP-tyrosine)
- And the aqueous solution contains free amino acids and Acid soluble DNP derivatives (diDNP-histidine, S-DNP cysteine, ⍺-DNP arginine, 𝜖-DNP-lysine, ⍺-DNP-lysine)
- Then dry the Ether extract and pass on ethanol-ligroin column followed by acetone-cyclohexane column to remove HCl and Ether.
- Determine the DNP product by colorimetric detection in comparison with standards.
Catalytic changes of Fructose 1,6-diphosphatase enzyme with DNFB
Pontremoli et al reported that 4 eq of DNFB per mole of the enzyme increases the catalytic activity of Fructose 1,6-diphosphatase by 3-4 times. Dinitrophenylated enzyme shows a change in optimum pH from 9.4 to 7.5. Also, the addition of DNFB results in a significant increase in Michaelis Constant for both substrates and metal ion activator. It is suggested that DNFB alters the allosteric site of the enzyme, thereby induces a change in protein conformation resulting in a change in activity.
Reagents
- 0.04 M Glycine buffer pH 9.4
- 0.04 M Triethanol Amine buffer pH 7.5
- 0.5 mM MnCl2
- 0.1 mM Fructose 1,6-diphosphatase
- DNFB
- 0.5mM TPN
- 5 µg Glucose 6-P dehydrogenase
- 5 µg Phosphogluco isomerase
Procedure
- All the above reagents should be mixed in a tube to a final volume of 1ml.
- Finally, the enzyme is added after all other reagents and mix gently.
- Incubate the reaction mixture for 10 min at 25 ℃.
- Stop the reaction by adding 0.1 ml 5N H2SO4.
- Free phosphate (Pi )was measured and compared between reactions with Dinitrophenylated enzyme and Untreated enzyme.
References
- Sanger F. (1945). The free amino groups of insulin. The Biochemical journal, 39(5), 507–515. doi:10.1042/j0390507
- Lorusso, M., Marzo, M., Gatti, D. and Papa, S.( 1986), Effect of 2,4-dinitrofluorobenzene on the enzymatic properties of the b-c1 complex isolated from beef heart mitochondria, FEBS Letters, 195, doi: 10.1016/0014-5793(86)80181-8
- Pontremoli, B. Luppis, W. A. Wood, S. Traniello, and B. L. Horecker Fructose Diphosphatase from Rabbit Liver: II. Changes in catalytic properties induced by Dinitrofluorobenzene . J. Biol. Chem. 1965 240: 3464.






