
Question:
From the seven protein assays you offer, which one is best for estimating the amount of peptide in my peptide solution?
The Protein Man Says:
G-Biosciences does offer seven different protein assays, which can be easily separated in to one of three assay types:
- Dye Binding Assays
- Copper Ion Based Assays
- Test Strip Based Assays
Dye Binding Assays
CB-X and CB protein assays are dye binding protein assays that are based on the binding of protein molecules to Coomassie dye under acidic conditions. The binding of protein to the dye results in spectral shift, the color shifts from brown (Amax= 465nm) to blue (Amax= 610nm). The change in color density is read at 595nm and is proportional to protein concentration. The REd 660 protein assay is based on a single proprietary dye-metal complex reagent. The binding of protein to the dye-metal complex under acidic conditions results in a change of color from reddish-brown to green and this change in color density is proportional to protein concentration. The color change is a result of deprotonation of the dye-metal complex at low pH, which is facilitated by interactions with positively charged amino acid groups.
A key feature of the dye binding assays is that the basic amino acids; arginine, lysine and histidine play a role in the formation of dye-protein complexes color. Small proteins or peptides less than 3kDa and amino acids generally do not produce color changes.
Dye binding assays are not suitable for estimating the concentration of protein assays.
Test Strip Based Assays
This is in effect a chromatographic capture method where the flat surface of the test strip acts as the solid matrix or support. Protein solution is applied on a specific protein binding test strip by point of contact capillary action. Under a specific buffer condition, as the protein enters into the matrix of the test strip, it binds instantly and saturates as the protein solution diffuses into the test strip in a circular manner. A circular protein imprint is produced which is developed into visible protein spots with a protein specific dye. The diameter of the protein spot is proportional to protein concentration. Measuring the protein spot diameter with a measuring gauge, the amount of protein can be estimated.
A key feature of the test strip based assays is that it uses a protein binding dye to show the protein spots. Again the basic amino acids; arginine, lysine and histidine play a role in the formation of dye-protein complexes color. Small proteins or peptides less than 3kDa and amino acids generally do not produce color changes.
Test strip based assays are not suitable for estimating the concentration of protein assays.
Copper Ion Based Assays
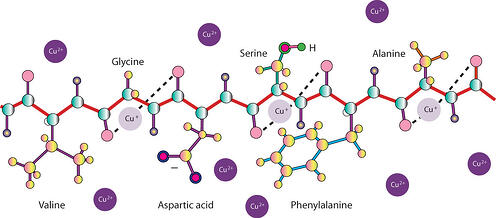
In the copper ion based protein assays, the protein solution is mixed with an alkaline solution of copper salt. Under alkaline conditions, cupric ions (Cu2+) chelate with the peptide bonds resulting in reduction of cupric (Cu2+) to cuprous ions (Cu+). If the alkaline copper is in excess over the amount of peptide bonds, some of the cupric ions (Cu2+) will remain unbound to the peptide bonds and are available for detection. Protein assays based on copper ions can be divided into two groups, assays that detect the reduced cuprous ions (Cu+) and that detect unbound cupric (Cu2+) ions.
The cuprous ions are detected either with bicinchoninic acid (BCA) or Folin Reagent (phosphomolybdic/ phosphotungstic acid). Cuprous ions (Cu+) reduction of BCA Reagent produces a purple color that can be read at 562nm. The amount of color produced is proportional to the amount of peptide bonds, i.e. size as well as the amount of protein/peptide.
The presence of tyrosine, tryptophan, cysteine, histidine and asparginine in protein contributes to additional reducing potential and enhances the amount of color produced. Hence, the amount of blue color produced is dependent on the composition of protein molecules. The reaction of cuprous ions (Cu+) with the bicinchoninic acid and color production is similar to that of Folin Reagent.
The NI Protein assay is based on the detection of unbound cupric ions, the protein solution is mixed with an amount of alkaline copper that is in excess over the amount of peptide bond. The unchelated cupric ions are detected with a color-producing reagent that reacts with cupric ions. The amount of color produced is inversely proportional to the amount of peptide bond.
In all cases the copper ions are binding to the protein or peptide, so are not dependent on basic amino acids to be present.
Copper ion based assays are the best option for detecting peptides, however, as with all assays, there is a limit of detection. So issues may arise with very small peptides or very dilute solutions.






